Dan Starr's Google Scholar
GW Gant Luxton's Google Scholar
Joint Starr & Luxton Publications
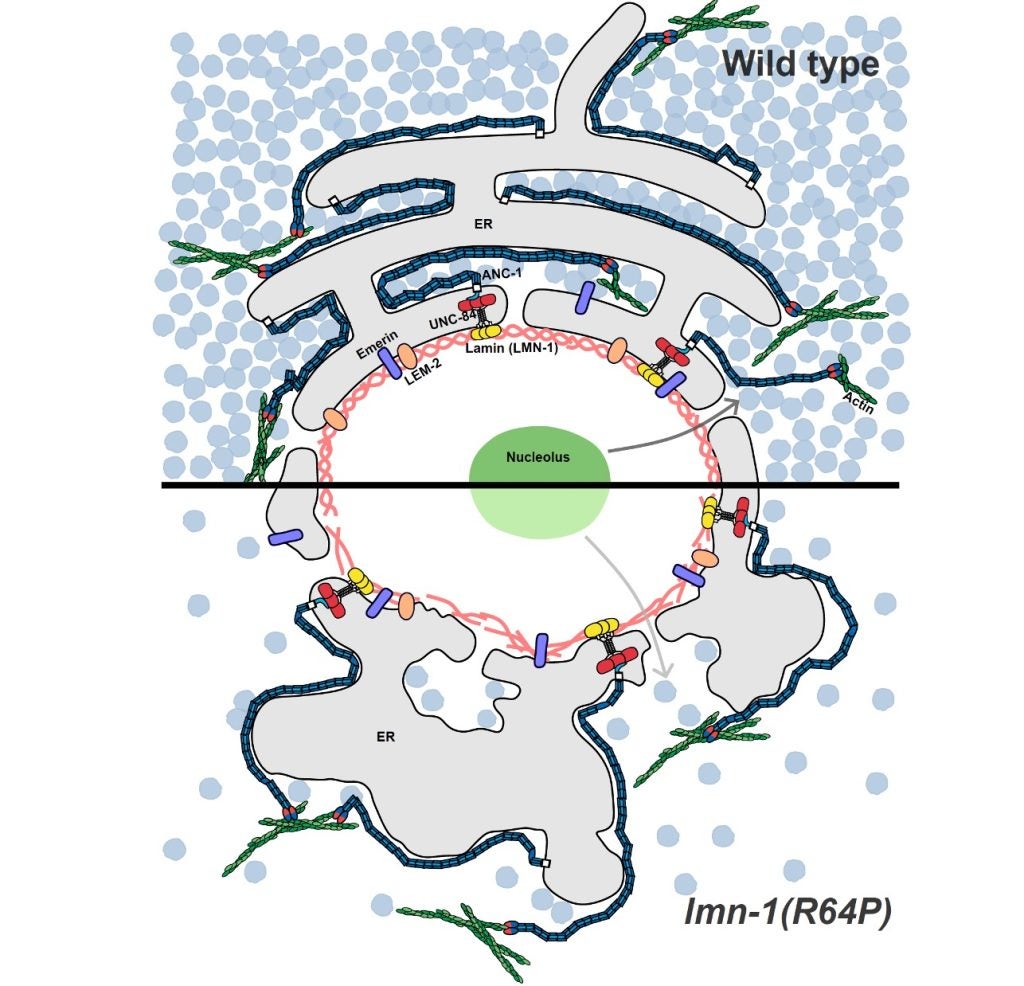
X Ding, S Kumari, E Gregory, DA Starr* and GWG Luxton* (2025), Muscular dystrophy-associated lamin variants disrupt cellular organization through a nucleolar-ribosomal axis controlling cytoplasmic macromolecular crowding. bioRxiv
Emery-Dreifuss muscular dystrophy (EDMD) arises from mutations in nuclear lamins or emerin. Current pathological models emphasize defects in nuclear mechanics and transcription regulation. Yet these models do not fully explain the complexity of EDMD or other laminopathies. Here, we uncover an emerging pathway linking nuclear lamina defects to the reorganization of cytoplasmic biophysics, revealing how nuclear dysfunction cascades throughout the cell. Using Caenorhabditis elegans EDMD models, we demonstrate that lamin mutations dramatically alter cytoplasmic organization, reducing macromolecular crowding and increasing diffusivity of 40 nm Genetically Encoded Multimeric (GEM) nanoparticles. These striking biophysical changes coincide with nuclear positioning defects and collapsed endoplasmic reticulum architecture, mirroring phenotypes associated with ribosome depletion. We propose a mechanism where mutations in the C. elegans lamin lmn-1 disrupt nucleolar density and ribosome biogenesis, creating a nucleolar-ribosomal axis that propagates defects from the nucleus to the cytoplasm. Genetic interactions between lmn-1 and ribosomes support this regulatory relationship. While individual depletion of other nuclear envelope proteins produces minimal effects, combined loss of the functionally redundant emerin ortholog emr-1 and LEM-domain protein lem-2 phenocopied lmn-1 mutants, demonstrating that cytoplasmic biophysical disruption lies at EDMD’s pathogenic core. Our findings establish a paradigm where nuclear lamina defects fundamentally rewire cellular biophysics through nucleolar-ribosomal dysfunction, opening transformative therapeutic avenues for treating laminopathies.
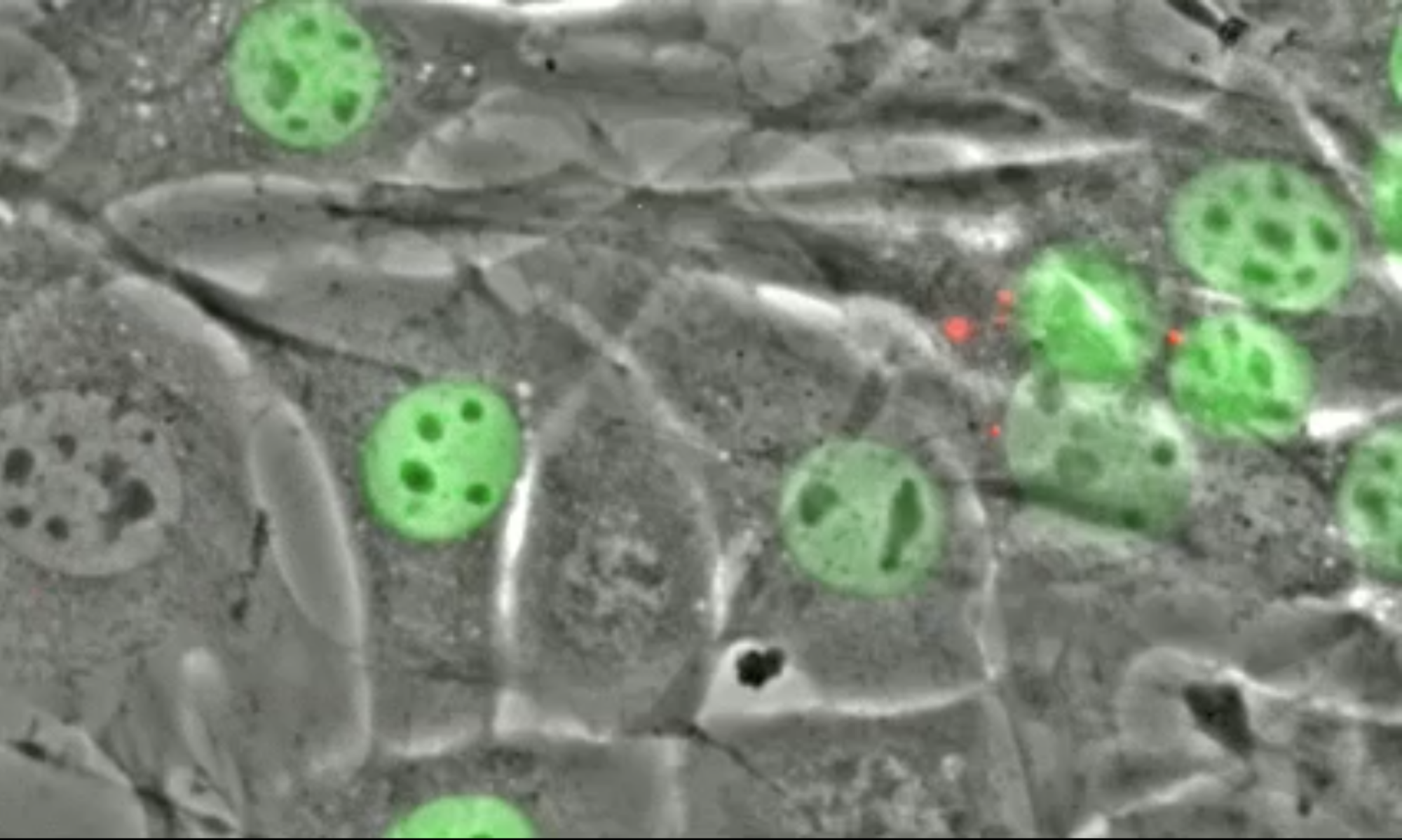
X Ding†, H Hao†, D Elnatan†, PN Alinaya, S Kalra, A Kaur, S Kumari, LJ Holt, GWG Luxton*, and DA Starr* (2025), Giant KASH proteins and ribosomes establish distinct cytoplasmic biophysical properties in vivo. Science Advances
Understanding how cells control their biophysical properties during development remains a fundamental challenge. While macromolecular crowding affects multiple cellular processes in single cells, its regulation in living animals remains poorly understood. Using genetically encoded multimeric nanoparticles for in vivo rheology, we found that Caenorhabditis elegans tissues maintain mesoscale properties that differ from those observed across diverse systems, including bacteria, yeast species, and cultured mammalian cells. We identified two conserved mechanisms controlling particle mobility: Ribosome concentration, a known regulator of cytoplasmic crowding, works in concert with a previously unknown function for the giant KASH (Klarsicht/ANC-1/SYNE homology) protein ANC-1 in providing structural constraints through associating with the endoplasmic reticulum. These findings reveal mechanisms by which tissues establish and maintain distinct mesoscale properties, with implications for understanding cellular organization across species.
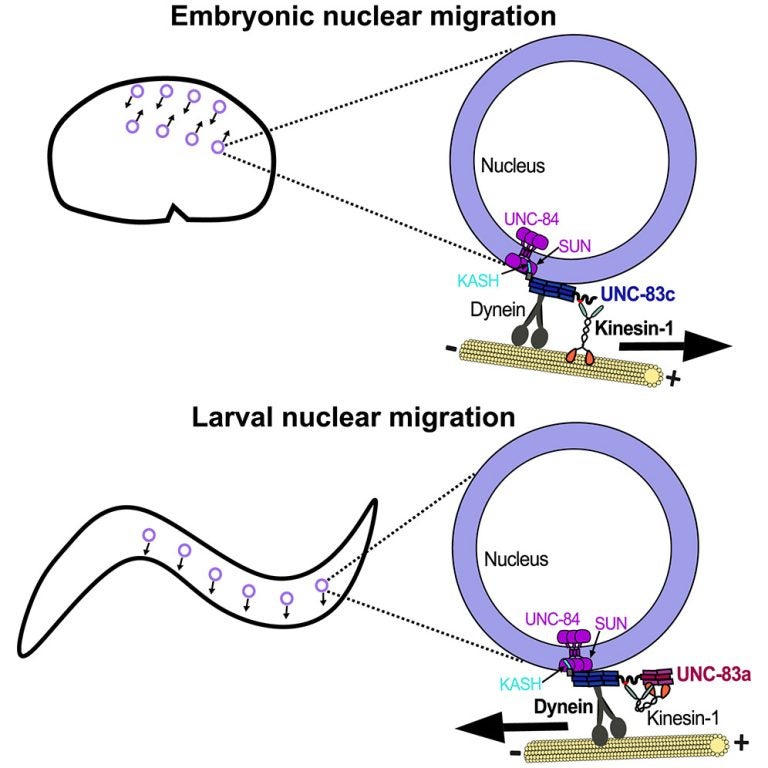
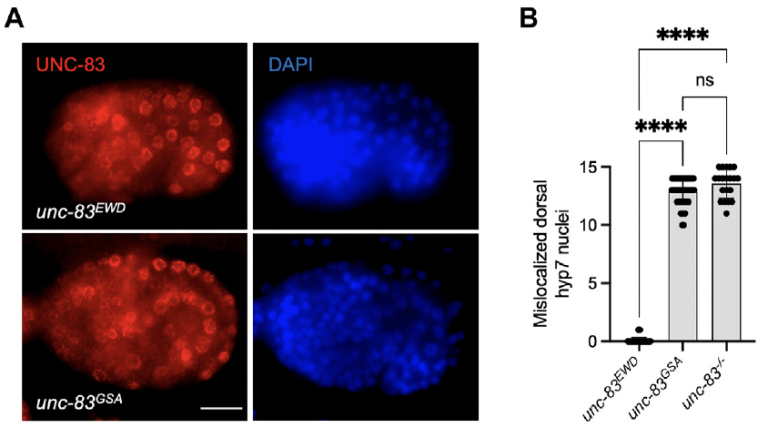
S Gümüşderelioğlu, N Sahabandu, D Elnatan, EF Gregory, K Chiba, S Niwa, GWG Luxton, RJ McKenney, and DA Starr (2025), The KASH protein UNC-83 differentially regulates kinesin-1 activity to control developmental stage-specific nuclear migration. Current Biology.
Nuclear migration plays a fundamental role in development, requiring precise spatiotemporal control of bidirectional movement through dynein and kinesin motors. Here, we uncover a differential isoform-dependent mechanism for developmental regulation of nuclear migration directionality. The nuclear envelope Klarsicht/ANC-1/Syne homology (KASH) protein UNC-83 in Caenorhabditis elegans exists in multiple isoforms that differentially control motor activity to achieve tissue-specific nuclear positioning. The shorter UNC-83c isoform promotes kinesin-1-dependent nuclear movement in embryonic hyp7 precursors, while longer UNC-83a/b isoforms facilitate dynein-mediated nuclear migration in larval P cells. We demonstrate that the UNC-83a-specific N-terminal domain functions as a kinesin-1 inhibitory module by directly binding the kinesin heavy chain (UNC-116). This interaction prevents kinesin-1 activation and reduces the protein’s affinity for kinesin light chain (KLC-2), allowing for dynein-mediated transport. By contrast, UNC-83c exhibits high-affinity binding to KLC-2, promoting kinesin-1 activation for plus-end-directed movement. AlphaFold structural predictions reveal that UNC-83 contains five spectrin-like repeats, with two located within the inhibitory N-terminal domain. Genetic analysis demonstrates that these spectrin-like repeats are essential for dynein-dependent P cell nuclear migration but dispensable for kinesin-1-dependent hyp7 migration. This isoform-specific inhibition, combined with differential affinity for KLC-2, establishes a mechanism for achieving directional control of nuclear positioning during development. Together, these interdisciplinary studies reveal how alternative isoforms of cargo adaptors can generate developmental stage-specific regulation of motor activity.
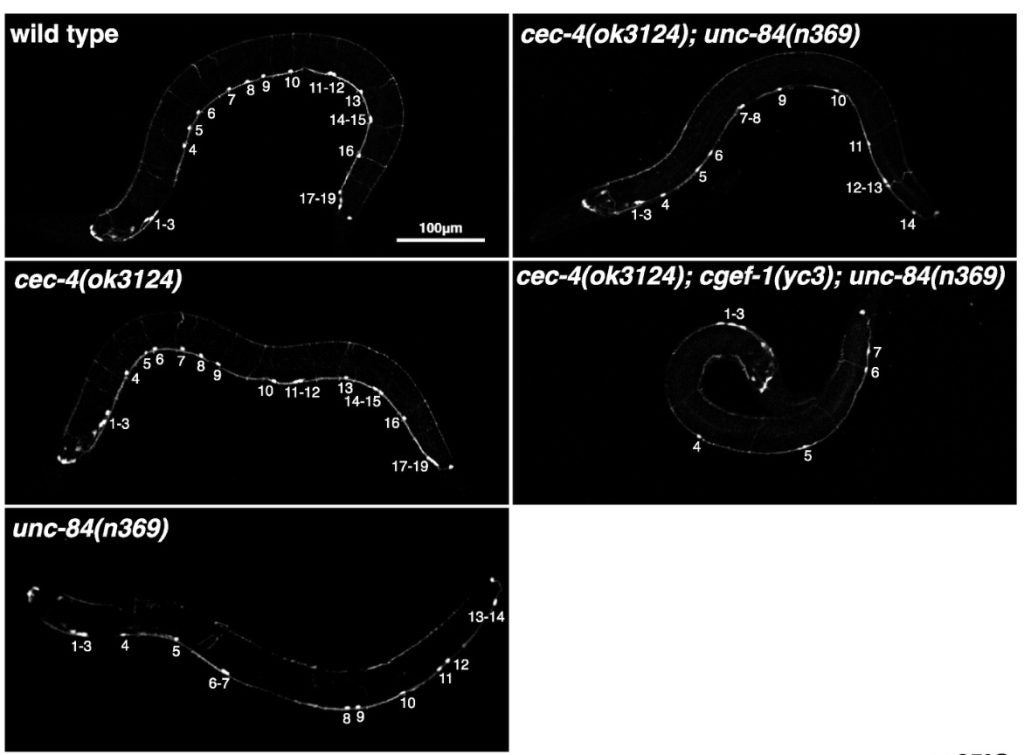
EF Gregory, GWG Luxton, DA Starr (2025), Nuclear deformability depends on H3K9-methylated heterochromatin anchorage to the nuclear periphery in Caenorhabditis elegans. Genetics.
Nuclei adjust their deformability while migrating through constrictions to enable structural changes and maintain nuclear integrity. The effect of heterochromatin anchored at the nucleoplasmic face of the inner nuclear membrane on nuclear morphology and deformability during in vivo nuclear migration through constricted spaces remains unclear. Here, we show that abolishing peripheral heterochromatin anchorage by eliminating CEC-4, a chromodomain protein that tethers H3K9-methylated chromatin to the nuclear periphery, disrupts constrained P-cell nuclear migration in Caenorhabditis elegans larvae in the absence of the established linker of nucleoskeleton and cytoskeleton (LINC) complex-dependent pathway. This effect was suppressed by mutations that stabilize the lamin LMN-1. CEC-4acts in parallel to an actin and CDC-42-based pathway. We also demonstrate the necessity for the chromatin methyltransferase MET-2 and the demethylase JMJD-1.2during P-cell nuclear migration in the absence of functional LINC complexes. We conclude that H3K9-methylated chromatin needs to be anchored to the nucleoplasmic face of the inner nuclear membrane to help facilitate nuclear migration through constricted spaces in vivo.
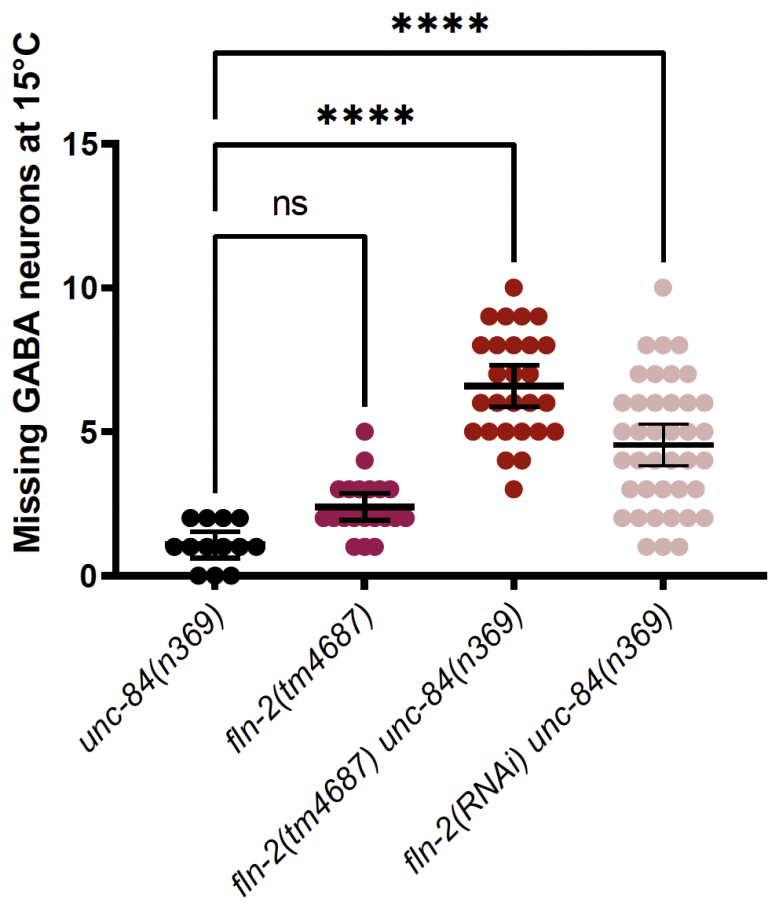
L Ma, J Kuhn, YT Chang, D Elnatan, GWG Luxton, and DA Starr (2024), FLN-2 functions in parallel to LINC complexes and Cdc42/actin pathways during P-cell nuclear migration through constricted spaces in Caenorhabditis elegans. Genetics
Nuclear migration through narrow constrictions is important for development, metastasis, and proinflammatory responses. Studies performed in tissue culture cells have implicated linker of nucleoskeleton and cytoskeleton (LINC) complexes, microtubule motors, the actin cytoskeleton, and nuclear envelope repair machinery as important mediators of nuclear movements through constricted spaces. However, little is understood about how these mechanisms operate to move nuclei in vivo. In Caenorhabditis elegans larvae, six pairs of hypodermal P cells migrate from lateral to ventral positions through a constricted space between the body wall muscles and the cuticle. P-cell nuclear migration is mediated in part by LINC complexes using a microtubule-based pathway and by an independent CDC-42/actin-based pathway. However, when both LINC complex and actin-based pathways are knocked out, many nuclei still migrate, suggesting the existence of additional pathways. Here, we show that FLN-2 functions in a third pathway to mediate P-cell nuclear migration. The predicted N-terminal actin-binding domain in FLN-2 that is found in canonical filamins is dispensable for FLN-2 function; this and structural predictions suggest that FLN-2 does not function as a filamin. The immunoglobulin-like repeats 4–8 of FLN-2 were necessary for P-cell nuclear migration. Furthermore, in the absence of the LINC complex component unc-84, fln-2 mutants had an increase in P-cell nuclear rupture. We conclude that FLN-2 functions to maintain the integrity of the nuclear envelope in parallel with the LINC complex and CDC-42/actin-based pathways to move P-cell nuclei through constricted spaces.
CT Liu, GWG Luxton, CC Sung, FM Wang, CN Chang, CF Hu (2023), Successful infliximab treatment for refractory COVID-19-associated multisystem inflammatory syndrome in two Taiwanese children: An experience from a medical center. Pediatrics and Neonatology.
Multiple inflammatory syndrome in children (MIS-C) is a rare, yet serious severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)-associated condition that typically presents 2–6 weeks after infection. MIS-C is believed to result from a delayed hyper-inflammatory reaction to SARS-CoV-2, which leads to an overwhelming cytokine storm, First-tier treatments for MIS-C include intravenous immunoglobulin (IVIG) and glucocorticoids as immune modulators; however, refractory cases may require the use of biologic agents as a second-tier intervention. The Taiwan Pediatric Association (TPA) recommends that MIS-C be treated first with an interleukin-1 receptor antagonist, anakinra, the use of which require prior approval from the Taiwan National Health Insurance system. To treat MIS-C cases that are unresponsive to standard first-tier treatments, we use the anti-tumor necrosis factor-alpha (TNF-α) antibody, infliximab, following clinical guidelines from the American College of Rheumatology (ACR). Here, we report two cases that benefited greatly from the prompt use of infliximab during refractory MIS-C management.
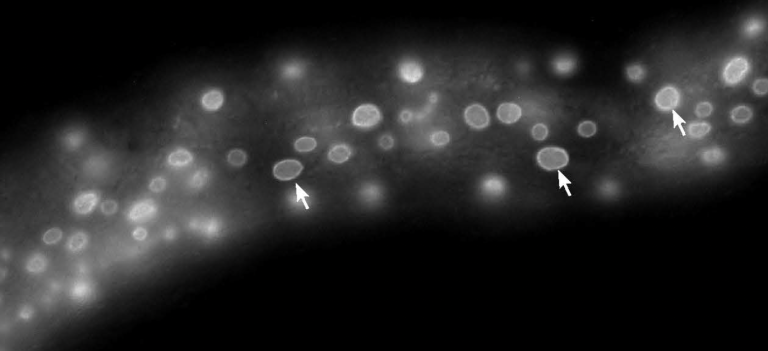
EF Gregory, JM Ragle, JD Ward, DA Starr (2023), Split-GFP lamin as a tool for studying C. elegans LMN-1 dynamics in vivo. microPublication Biology.
RM McGillivary, DA Starr*, GWG Luxton* (2023), Building and breaking mechanical bridges between the nucleus and cytoskeleton: Regulation of LINC complex assembly and disassembly. Current Opinion in Cell Biology. (*Equal contribution)
The nucleus is physically coupled to the cytoskeleton through LINC complexes, macromolecular bridges composed of SUN and KASH proteins that span the nuclear envelope. LINC complexes are involved in a wide variety of critical cellular processes. For these processes to occur, cells regulate the composition, assembly, and disassembly of LINC complexes. Here we discuss recent studies on the regulation of the SUN-KASH interaction that forms the core of the LINC complex. These new findings encompass the stages of LINC complex assembly, from the formation of SUN-KASH heterooligomers to higher-order assemblies of LINC complexes. There is also new work on how components of the LINC complex are selectively dismantled, particularly by proteasomal degradation. It is becoming increasingly clear that LINC complexes are subject to multiple layers of regulation.
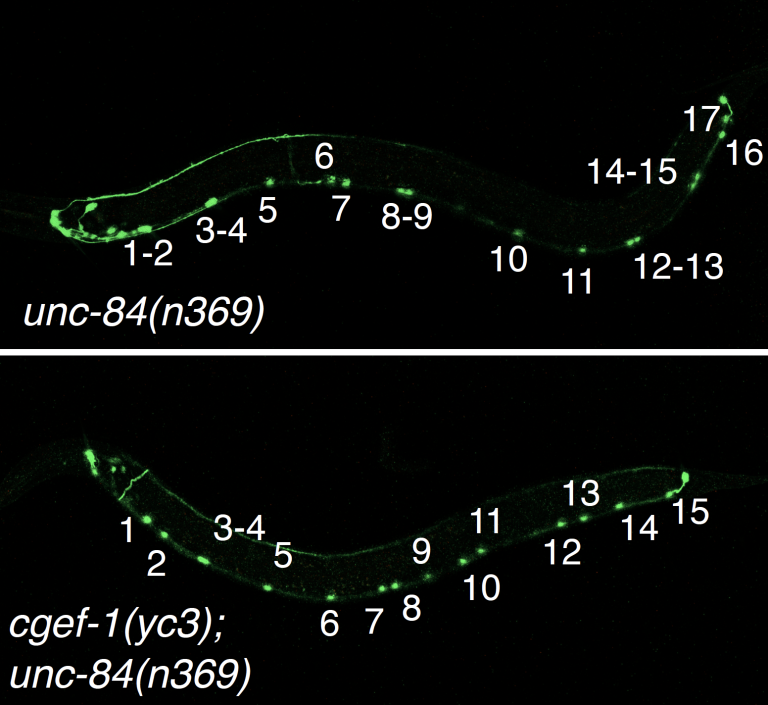
J Ho, LA Guerrero, DE Libuda, GWG Luxton, and DA Starr (2023), Actin and CDC-42 contribute to nuclear migration through constricted spaces in C. elegans. Development.
Successful nuclear migration through constricted spaces between cells or in the extracellular matrix relies on the ability of the nucleus to deform. Little is known about how this takes place in vivo. We have studied confined nuclear migration in Caenorhabditis elegans larval P cells, which is mediated by the LINC complex to pull nuclei towards the minus ends of microtubules. Null mutations of the LINC component unc-84 lead to a temperature-dependent phenotype, suggesting a parallel pathway for P-cell nuclear migration. A forward genetic screen for enhancers of unc-84 identified cgef-1 (CDC-42 guanine nucleotide exchange factor). Knockdown of CDC-42 in the absence of the LINC complex led to a P-cell nuclear migration defect. Expression of constitutively active CDC-42 partially rescued nuclear migration in cgef-1; unc-84 double mutants, suggesting that CDC-42 functions downstream of CGEF-1. The Arp2/3 complex and non-muscle myosin II (NMY-2) were also found to function parallel to the LINC pathway. In our model, CGEF-1 activates CDC-42, which induces actin polymerization through the Arp2/3 complex to deform the nucleus during nuclear migration, and NMY-2 helps to push the nucleus through confined spaces.
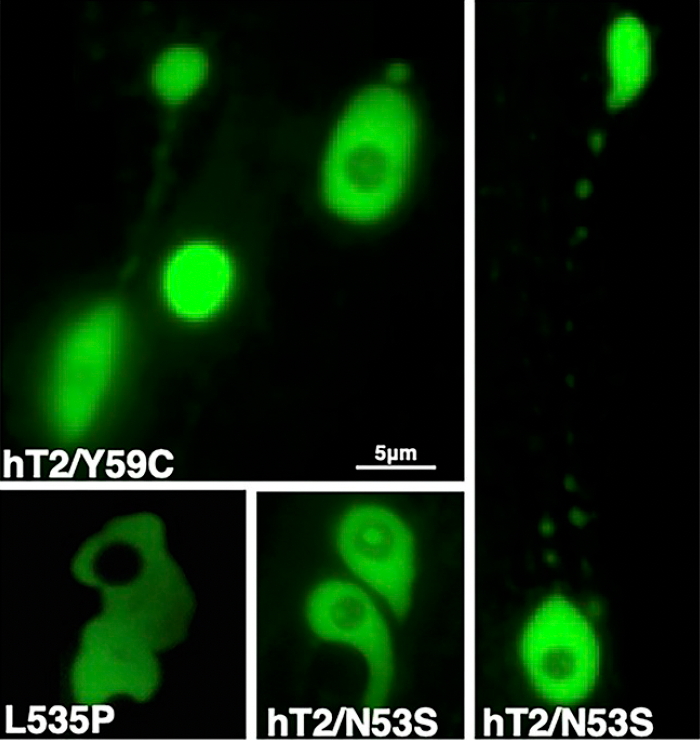
EF Gregory, S Kalra, T Brock, G Bonne, GWG Luxton, C Hopkins, and DA Starr (2023), Caenorhabditis elegans models for striated muscle disorders caused by missense variants of human LMNA. PLoS Genetics.
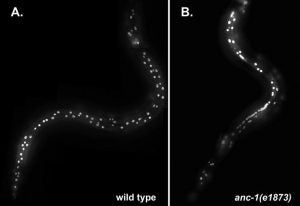
Striated muscle laminopathies caused by missense mutations in the nuclear lamin gene LMNA are characterized by cardiac dysfunction and often skeletal muscle defects. Attempts to predict which LMNA variants are pathogenic and to understand their physiological effects lag behind variant discovery. We created Caenorhabditis elegans models for striated muscle laminopathies by introducing pathogenic human LMNA variants and variants of unknown significance at conserved residues within the lmn-1 gene. Severe missense variants reduced fertility and/or motility in C. elegans. Nuclear morphology defects were evident in the hypodermal nuclei of many lamin variant strains, indicating a loss of nuclear envelope integrity. Phenotypic severity varied within the two classes of missense mutations involved in striated muscle disease, but overall, variants associated with both skeletal and cardiac muscle defects in humans lead to more severe phenotypes in our model than variants predicted to disrupt cardiac function alone. We also identified a separation of function allele, lmn-1(R204W), that exhibited normal viability and swimming behavior but had a severe nuclear migration defect. Thus, we established C. elegans avatars for striated muscle laminopathies and identified LMNA variants that offer insight into lamin mechanisms during normal development.
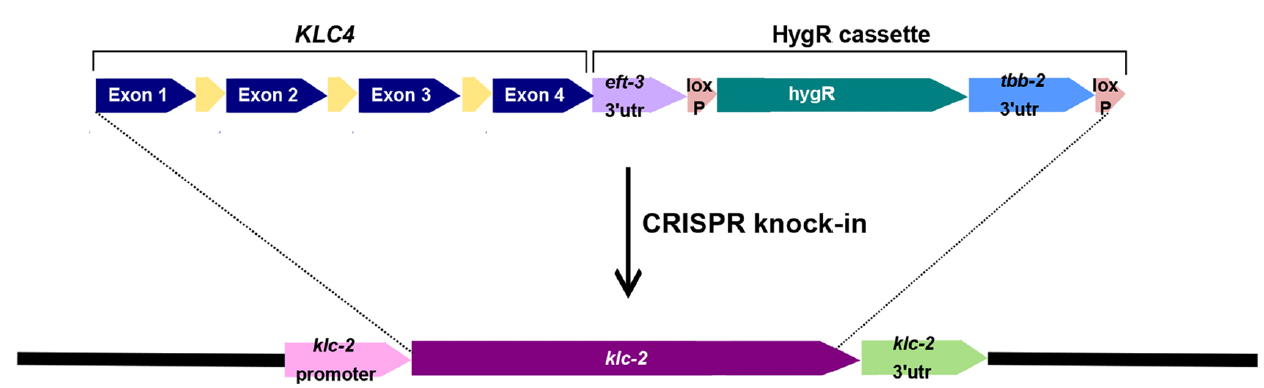
S Gümüşderelioğlu, L Resch, T Brock, GWG Luxton, H Cope, QKG Tan, C Hopkins, DA Starr (2023), A humanized Caenorhabditis elegans model of hereditary spastic paraplegia-associated variants in KLC4. Disease Models and Mechanisms.
Hereditary spastic paraplegia (HSP) is a group of degenerative neurological disorders. We identified a variant in human kinesin light chain 4 (KLC4) that is suspected to be associated with autosomal-dominant HSP. How this and other variants relate to pathologies is unknown. We created a humanized Caenorhabditis elegans model in which klc-2 was replaced by human KLC4 (referred to as hKLC4) and assessed the extent to which hKLC4 retained function in the worm. We observed a slight decrease in motility but no nuclear migration defects in the humanized worms, suggesting that hKLC4 retains much of the function of klc-2. Five hKLC4 variants were introduced into the humanized model. The clinical variant led to early lethality, with significant defects in nuclear migration when homozygous and a weak nuclear migration defect when heterozygous, possibly correlating with the clinical finding of late-onset HSP when the proband was heterozygous. Thus, we were able to establish humanized C. elegans as an animal model for HSP and to use it to test the significance of five variants of uncertain significance in the human gene KLC4.
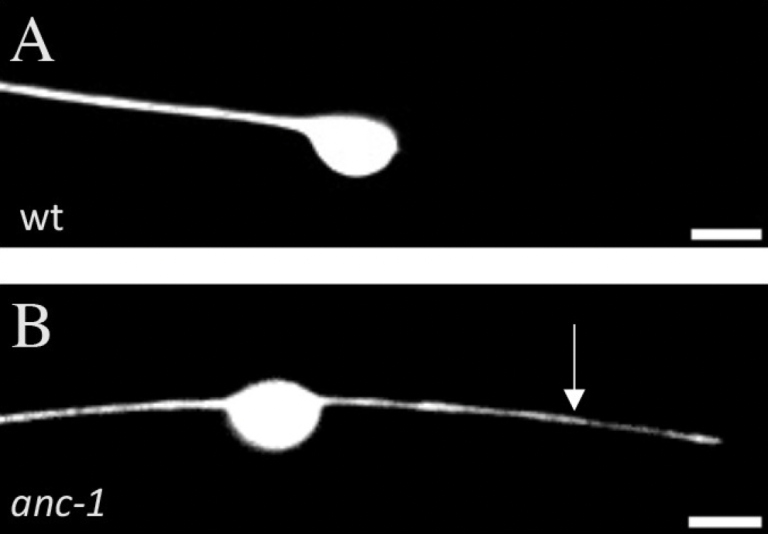
NC Fischer, V Friedman, MA Martinez-Reyes, H Hao, TA Chowdhury, DA Starr, CC Quinn (2022), The ANC-1 (Nesprin-1/2) organelle-anchoring protein functions through mitochondria to polarize axon growth in response to SLT-1. PLoS Genetics.
A family of giant KASH proteins, including C. elegans ANC-1 and mammalian Nesprin-1 and -2, are involved in organelle anchoring and are associated with multiple neurodevelopmental disorders including autism, bipolar disorder, and schizophrenia. However, little is known about how these proteins function in neurons. Moreover, the role of organelle anchoring in axon development is poorly understood. Here, we report that ANC-1 functions with the SLT-1 extracellular guidance cue to polarize ALM axon growth. This role for ANC-1 is specific to its longer ANC-1A and ANC-1C isoforms, suggesting that it is mechanistically distinct from previously described roles for ANC-1. We find that ANC-1 is required for the localization of a cluster of mitochondria to the base of the proximal axon. Furthermore, genetic and pharmacological studies indicate that ANC-1 functions with mitochondria to promote polarization of ALM axon growth. These observations reveal a mechanism whereby ANC-1 functions through mitochondria to polarize axon growth in response to SLT-1.
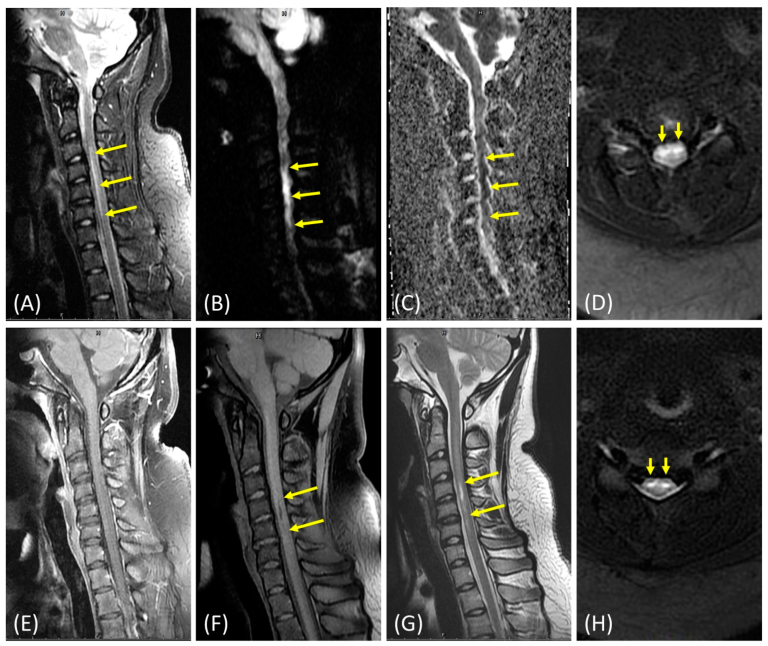
CC Hu, YY Yang, GWG Luxton, YP Lin, KS Hung, CF Hu (2022), The Role of Folate Deficiency as a Potential Risk Factor for Nontraumatic Anterior Spinal Artery Syndrome in an Adolescent Girl. Brain Sciences.
Nuclear positioning is important for the functionality of many cell types and is mediated by interactions of cytoskeletal elements and nucleoskeleton proteins. Nesprin proteins, part of the linker of nucleoskeleton and cytoskeleton (LINC) complex, have been shown to participate in nuclear positioning in multiple cell types. Outer hair cells (OHCs) in the inner ear are specialized sensory epithelial cells that utilize somatic electromotility to amplify auditory signals in the cochlea. Recently, Nesprin-4 (encoded by Syne4) was shown to play a crucial role in nuclear positioning in OHCs. Syne4 deficiency in humans and mice leads to mislocalization of the OHC nuclei and cell death resulting in deafness. However, it is unknown how Nesprin-4 mediates the position of the nucleus, and which other molecular components are involved in this process. Here, we show that the interaction of Nesprin-4 and the microtubule motor kinesin-1 is mediated by a conserved 4 amino-acid motif. Using in vivo AAV gene delivery, we show that this interaction is critical for nuclear positioning and hearing in mice. Nuclear mislocalization and cell death of OHCs coincide with the onset of hearing and electromotility and are solely restricted to outer, but not inner, hair cells. Likewise, the C. elegans functional homolog of Nesprin-4, UNC-83, uses a similar motif to mediate interactions between migrating nuclei and kinesin-1. Overall, our results suggest that OHCs require unique cellular machinery for proper nuclear positioning at the onset of electromotility. This machinery relies on the interaction between Nesprin-4 and kinesin-1 motors supporting a microtubule cargo model for nuclear positioning.
S Taiber, O Gozlan, R Cohen, LR Andrade, EF Gregory, DA Starr, Y Moran, R Hipp, MW Kelley, U Manor, D Sprinzak, KB Avraham (2022), A Nesprin-4/kinesin-1 cargo model for nuclear positioning in cochlear outer hair cells. Frontiers in Cell and Developmental Biology.
Nuclear positioning is important for the functionality of many cell types and is mediated by interactions of cytoskeletal elements and nucleoskeleton proteins. Nesprin proteins, part of the linker of nucleoskeleton and cytoskeleton (LINC) complex, have been shown to participate in nuclear positioning in multiple cell types. Outer hair cells (OHCs) in the inner ear are specialized sensory epithelial cells that utilize somatic electromotility to amplify auditory signals in the cochlea. Recently, Nesprin-4 (encoded by Syne4) was shown to play a crucial role in nuclear positioning in OHCs. Syne4 deficiency in humans and mice leads to mislocalization of the OHC nuclei and cell death resulting in deafness. However, it is unknown how Nesprin-4 mediates the position of the nucleus, and which other molecular components are involved in this process. Here, we show that the interaction of Nesprin-4 and the microtubule motor kinesin-1 is mediated by a conserved 4 amino-acid motif. Using in vivo AAV gene delivery, we show that this interaction is critical for nuclear positioning and hearing in mice. Nuclear mislocalization and cell death of OHCs coincide with the onset of hearing and electromotility and are solely restricted to outer, but not inner, hair cells. Likewise, the C. elegans functional homolog of Nesprin-4, UNC-83, uses a similar motif to mediate interactions between migrating nuclei and kinesin-1. Overall, our results suggest that OHCs require unique cellular machinery for proper nuclear positioning at the onset of electromotility. This machinery relies on the interaction between Nesprin-4 and kinesin-1 motors supporting a microtubule cargo model for nuclear positioning.
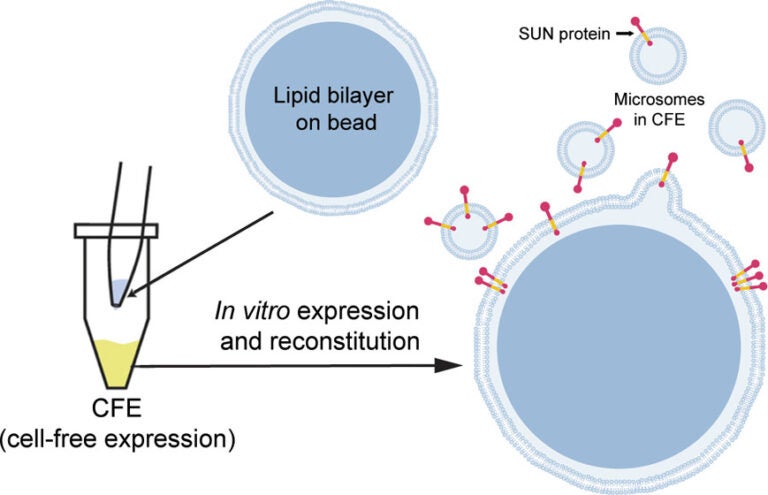
S Majumder, YY Hsu, H Moghimianavval, M Andreas, TW Giessen, GWG Luxton, AP Liu (2022), In Vitro Synthesis and Reconstitution Using Mammalian Cell-Free Lysates Enables the Systematic Study of the Regulation of LINC Complex Assembly. Biochemistry.
Understanding the structure and structure–function relationships of membrane proteins is a fundamental problem in biomedical research. Given the difficulties inherent to performing mechanistic biochemical and biophysical studies of membrane proteins in vitro, we previously developed a facile HeLa cell-based cell-free expression (CFE) system that enables the efficient reconstitution of full-length (FL) functional inner nuclear membrane Sad1/UNC-84 (SUN) proteins (i.e., SUN1 and SUN2) in supported lipid bilayers. Here, we provide evidence that suggests that the reconstitution of CFE-synthesized FL membrane proteins in supported lipid bilayers occurs primarily through the fusion of endoplasmic reticulum-derived microsomes present within our CFE reactions with our supported lipid bilayers. In addition, we demonstrate the ease with which our synthetic biology platform can be used to investigate the impact of the chemical environment on the ability of CFE-synthesized FL SUN proteins reconstituted in supported lipid bilayers to interact with the luminal domain of the KASH protein nesprin-2. Moreover, we use our platform to study the molecular requirements for the homo- and heterotypic interactions between SUN1 and SUN2. Finally, we show that our platform can be used to simultaneously reconstitute three different CFE-synthesized FL membrane proteins in a single supported lipid bilayer. Overall, these results establish our HeLa cell-based CFE and supported lipid bilayer reconstitution platform as a powerful tool for performing mechanistic dissections of the oligomerization and function of FL membrane proteins in vitro. While our platform is not a substitute for cell-based studies, it does provide important mechanistic insights into the biology of difficult-to-study membrane proteins.
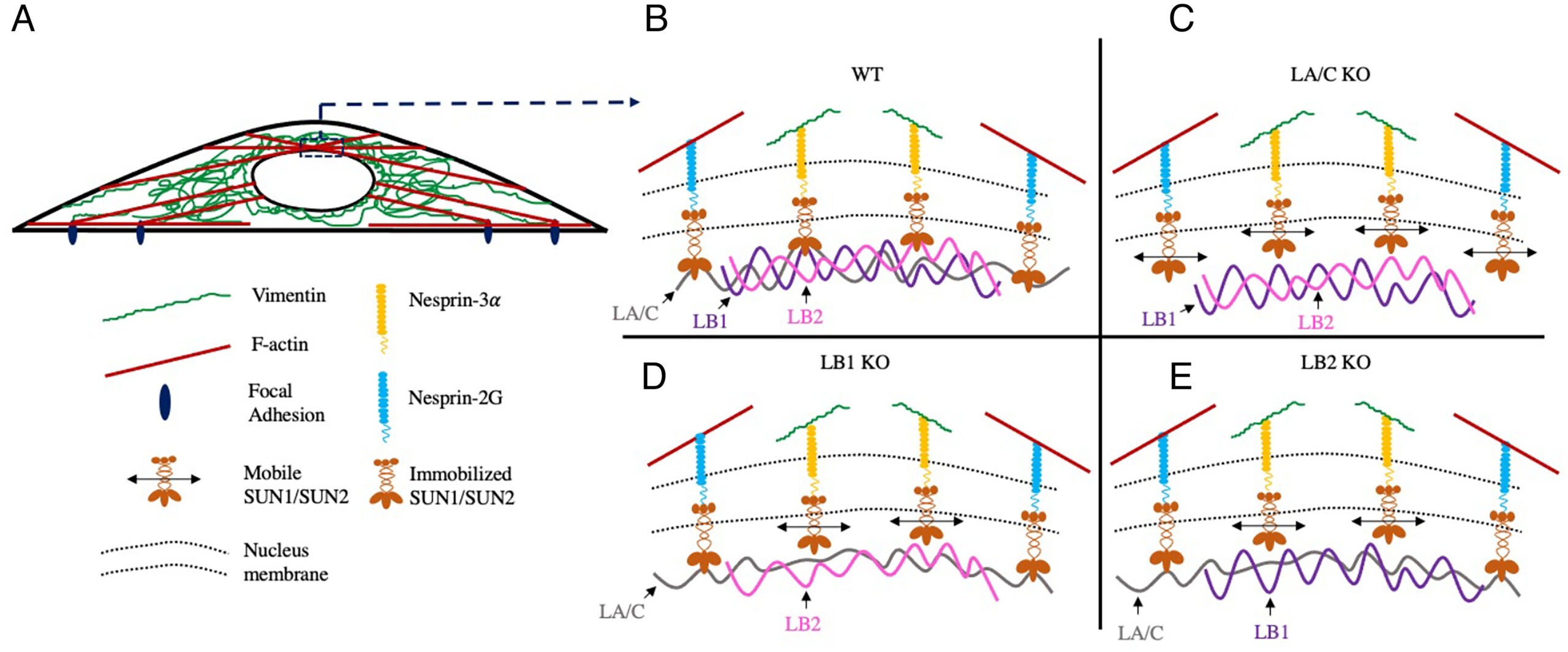
A Vahabikashi, S Sivagurunathan, FA Sadsad Nicdao, YL Han, CY Park, M Kittisopikul, X Wong, JR Tran, GG Gundersen, KL Reddy, GWG Luxton, M Guo, JJ Fredberg, Y Zheng, SA Adam, RD Goldman (2022), Nuclear lamin isoforms differentially contribute to LINC complex-dependent nucleocytoskeletal coupling and whole-cell mechanics. PNAS.
The ability of a cell to regulate its mechanical properties is central to its function. Emerging evidence suggests that interactions between the cell nucleus and cytoskeleton influence cell mechanics through poorly understood mechanisms. Here we conduct quantitative confocal imaging to show that the loss of A-type lamins tends to increase nuclear and cellular volume while the loss of B-type lamins behaves in the opposite manner. We use fluorescence recovery after photobleaching, atomic force microscopy, optical tweezer microrheology, and traction force microscopy to demonstrate that A-type lamins engage with both F-actin and vimentin intermediate filaments (VIFs) through the linker of nucleoskeleton and cytoskeleton (LINC) complexes to modulate cortical and cytoplasmic stiffness as well as cellular contractility in mouse embryonic fibroblasts (MEFs). In contrast, we show that B-type lamins predominantly interact with VIFs through LINC complexes to regulate cytoplasmic stiffness and contractility. We then propose a physical model mediated by the lamin–LINC complex that explains these distinct mechanical phenotypes (mechanophenotypes). To verify this model, we use dominant negative constructs and RNA interference to disrupt the LINC complexes that facilitate the interaction of the nucleus with the F-actin and VIF cytoskeletons and show that the loss of these elements results in mechanophenotypes like those observed in MEFs that lack A- or B-type lamin isoforms. Finally, we demonstrate that the loss of each lamin isoform softens the cell nucleus and enhances constricted cell migration but in turn increases migration-induced DNA damage. Together, our findings uncover distinctive roles for each of the four major lamin isoforms in maintaining nucleocytoskeletal interactions and cellular mechanics.
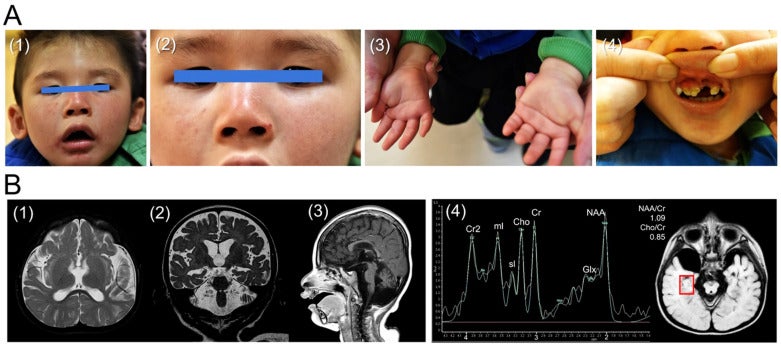
CM Lin, JH Yang, HJ Lee, YP Lin, LP Tsai, CS Hsu, GWG Luxton*, CF Hu* (2021), Whole Exome Sequencing Identifies a Novel Homozygous Missense Mutation in the CSB Protein-Encoding ERCC6 Gene in a Taiwanese Boy with Cockayne Syndrome. Life. (* Equal contribution)
Cockayne syndrome (CS) is a rare form of dwarfism that is characterized by progressive premature aging. CS is typically caused by mutations in the excision repair cross-complementing protein group 6 (ERCC6) gene that encodes the CS group B (CSB) protein. Using whole exome sequencing, we recently identified a novel homozygous missense mutation (Leu536Trp) in CSB in a Taiwanese boy with CS. Since the current database (Varsome) interprets this variant as likely pathogenic, we utilized a bioinformatic tool to investigate the impact of Leu536Trp as well as two other variants (Arg453Ter, Asp532Gly) in similar articles on the CSB protein structure stability. We used iterative threading assembly refinement (I-TASSER) to generate a predictive 3D structure of CSB. We calculated the change of mutation energy after residues substitution on the protein stability using I-TASSER as well as the artificial intelligence program Alphafold. The Asp532Gly variant destabilized both modeled structures, while the Leu536Trp variant showed no effect on I-TASSER’s model but destabilized the Alphafold’s modeled structure. We propose here the first case of CS associated with a novel homozygous missense mutation (Leu536Trp) in CSB. Furthermore, we suggest that the Asp532Gly and Leu536Trp variants are both pathogenic after bioinformatic analysis of protein stability.
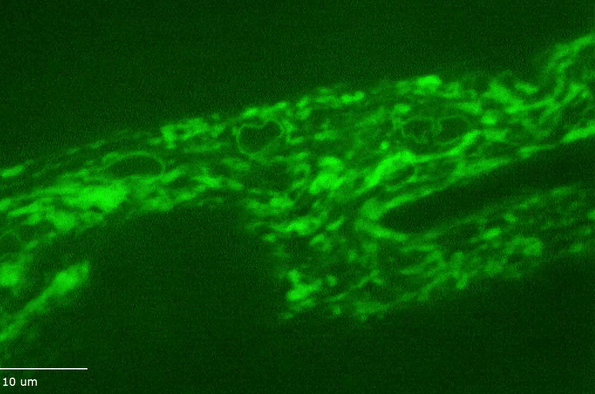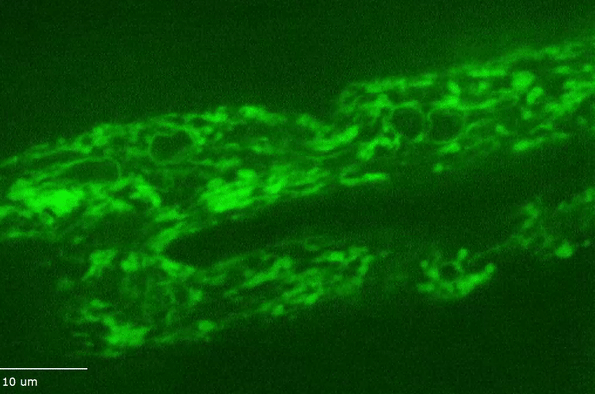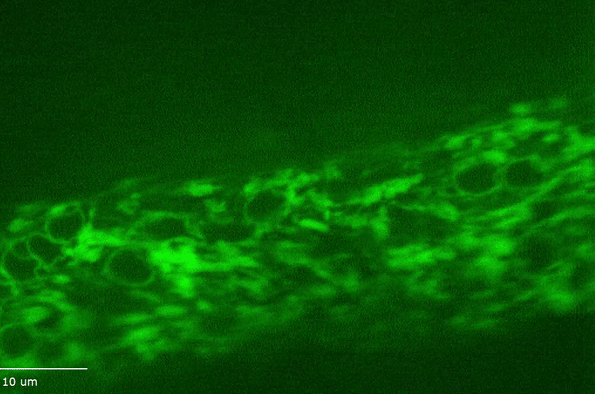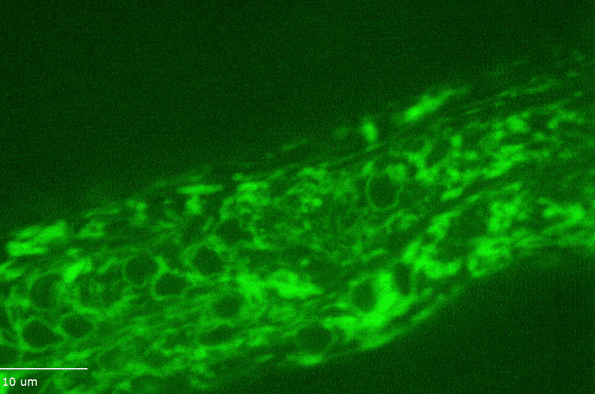
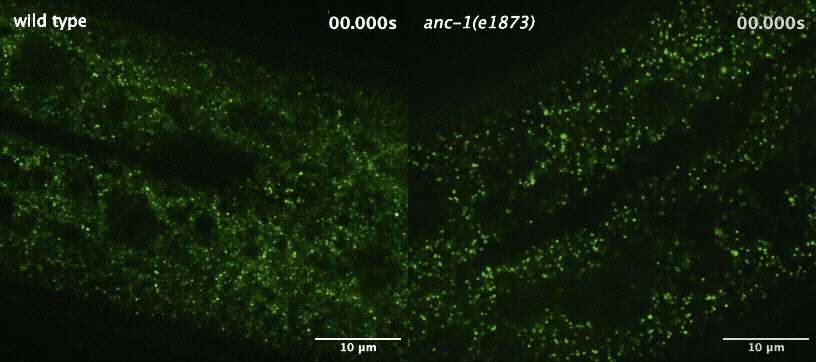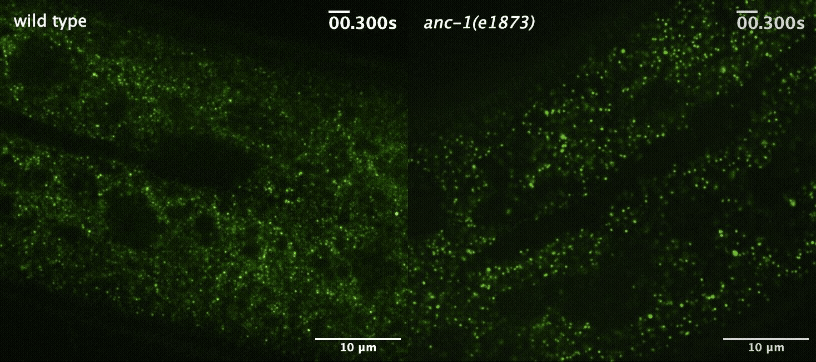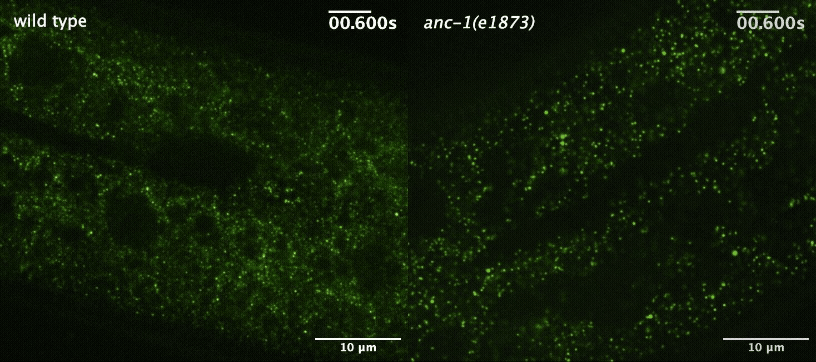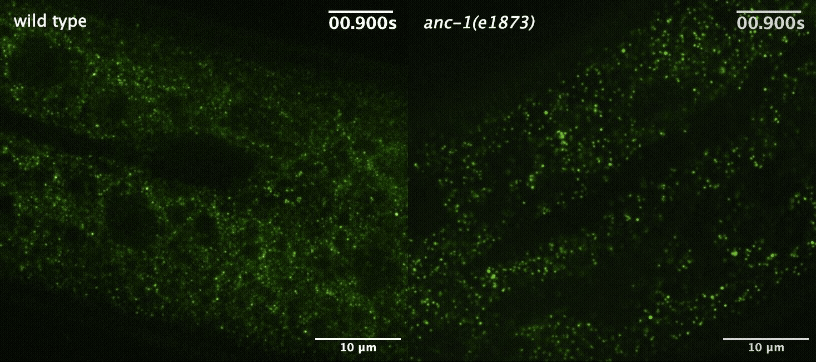
H Hao, S Kalra, LE Jameson, LA Guerrero, NE Cain, J Bolivar and DA Starr (2021), The Nesprin-1/-2 ortholog ANC-1 regulates organelle positioning in C. elegans independently from its KASH or actin-binding domains. eLife.
KASH proteins in the outer nuclear membrane comprise the cytoplasmic half of LINC complexes that connect nuclei to the cytoskeleton. Caenorhabditis elegans ANC-1, an ortholog of Nesprin-1/2, contains actin-binding and KASH domains at opposite ends of a long spectrin-like region. Deletion of either the KASH or calponin homology (CH) domains does not completely disrupt nuclear positioning, suggesting neither KASH nor CH domains are essential. Deletions in the spectrin-like region of ANC-1 led to significant defects, but only recapitulated the null phenotype in combination with mutations in the trans-membrane span. In anc-1 mutants, the ER was unanchored, moving throughout the cytoplasm, and often fragmented. The data presented here support a cytoplasmic integrity model where ANC-1 localizes to the ER membrane and extends into the cytoplasm to position nuclei, ER, mitochondria, and likely other organelles in place.
Pre — 2021 Starr Publications Google Scholar
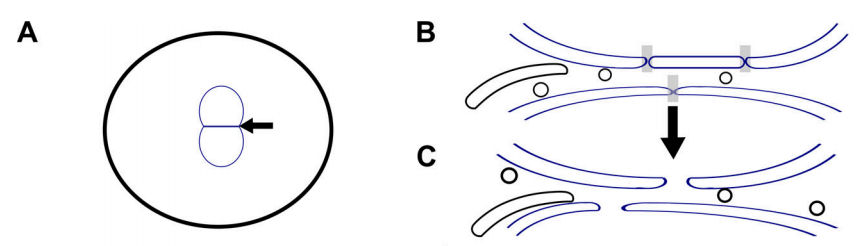
L Ma and DA Starr (2020), Membrane fusion drives pronuclear meeting in the one-cell embryo. The Journal of Cell Biology invited perspective.
The mechanisms that control how the two parental pronuclei fuse in the first mitosis of the embryo are poorly understood. In this issue, Rahman et al. (2020. J. Cell Biol.) found that membrane fusion between pronuclear envelopes, followed by fenestration, promotes pronuclear fusion.
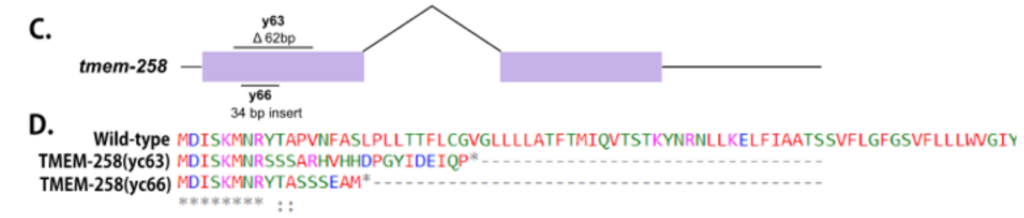
EF Gregory and DA Starr (2020), tmem-258 is dispensable for both nuclear anchorage and migration in C. elegans. microPublication Biology.
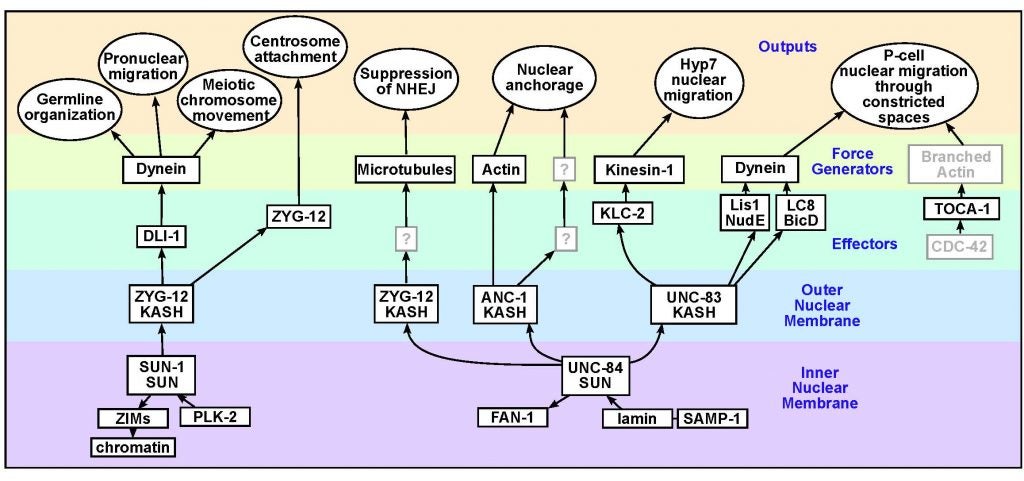
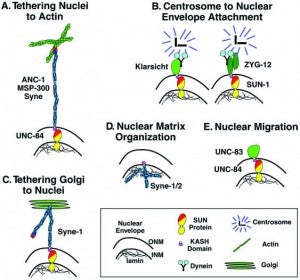
DA Starr (2019), A network of nuclear envelope proteins and cytoskeletal force generators mediates movements of and within nuclei throughout C. elegans development. Experimental Biology and Medicine.
Nuclear migration and anchorage, together referred to as nuclear positioning, are central to many cellular and developmental events. Nuclear positioning is mediated by a conserved network of nuclear envelope proteins that interacts with force generators in the cytoskeleton. At the heart of this network are LINC (linker of nucleoskeleton and cytoskeleton) complexes made of SUN (Sad1 and UNC-84) proteins at the inner nuclear membrane and KASH (Klarsicht, ANC-1, and Syne homology) proteins in the outer nuclear membrane. LINC complexes span the nuclear envelope, maintain nuclear envelope architecture, designate the surface of nuclei distinctly from the contiguous ER, and were instrumental in the early evolution of eukaryotes. LINC complexes interact with lamins in the nucleus and with various cytoplasmic KASH effectors from the surface of nuclei. These effectors regulate the cytoskeleton, leading to a variety of cellular outputs including pronuclear migration, nuclear migration through constricted spaces, nuclear anchorage, centrosome attachment to nuclei, meiotic chromosome movements, and DNA damage repair. How LINC complexes are regulated and how they function are reviewed here. The focus is on recent studies elucidating the best-understood network of LINC complexes, those used throughout C. elegans development.
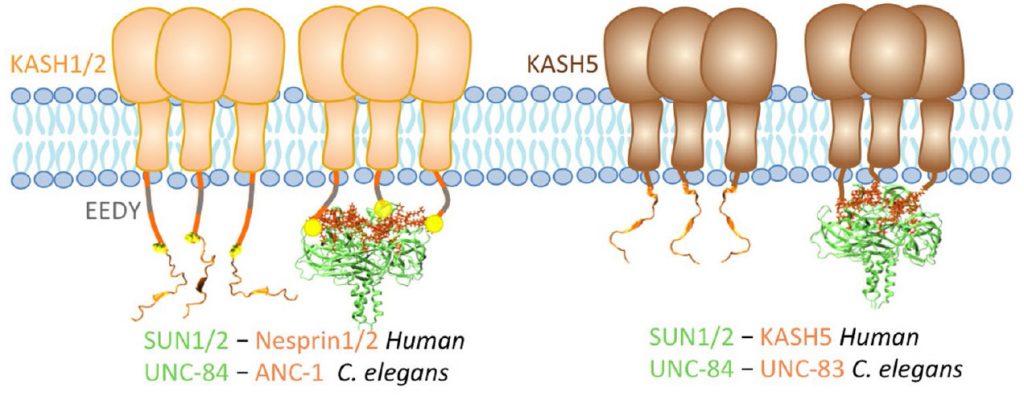
Z Jahed, H Hao, V Thakkar, UT Vu, A Valdez, A Ratish, C Tolentino, SCJ Kim, D Fadavi, DA Starr, MRK Mofrad (2019), Role of KASH lengths in the regulation of LINC complexes. Molecular Biology of the Cell.
The linker of the nucleoskeleton and cytoskeleton (LINC) complex is formed by the conserved interactions between Sad-1 and UNC-84 (SUN) and Klarsicht, ANC-1, SYNE homology (KASH) domain proteins, providing a physical coupling between the nucleoskeleton and cytoskeleton that mediates the transfer of physical forces across the nuclear envelope. The LINC complex can perform distinct cellular functions by pairing various KASH domain proteins with the same SUN domain protein. For example, in Caenorhabditis elegans, SUN protein UNC-84 binds to two KASH proteins UNC-83 and ANC-1 to mediate nuclear migration and anchorage, respectively. In addition to distinct cytoplasmic domains, the luminal KASH domain also varies among KASH domain proteins of distinct functions. In this study, we combined in vivo C. elegans genetics and in silico molecular dynamics simulations to understand the relation between the length and amino acid composition of the luminal KASH domain, and the function of the SUN–KASH complex. We show that longer KASH domains can withstand and transfer higher forces and interact with the membrane through a conserved membrane proximal EEDY domain that is unique to longer KASH domains. In agreement with our models, our in vivo results show that swapping the KASH domains of ANC-1 and UNC-83, or shortening the KASH domain of ANC-1, both result in a nuclear anchorage defect in C. elegans.
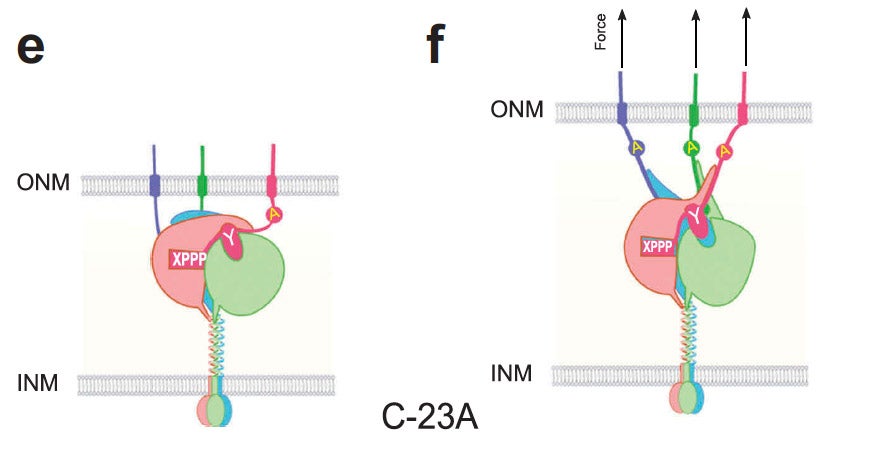
H Hao, DA Starr (2019), SUN/KASH interactions facilitate force transmission across the nuclear envelope. Nucleus.
LINC complexes (Linker of Nucleoskeleton and Cytoskeleton), consisting of inner nuclear membrane SUN (Sad1, UNC-84) proteins and outer nuclear membrane KASH (Klarsicht, ANC-1, and Syne Homology) proteins, are essential for nuclear positioning, cell migration and chromosome dynamics. To test the in vivo functions of conserved interfaces revealed by crystal structures, Cain et al used a combination of Caenorhabditis elegans genetics, imaging in cultured NIH 3T3 fibroblasts, and Molecular Dynamic simulations, to study SUN-KASH interactions. Conserved aromatic residues at the -7 position of the C-termini of KASH proteins and conserved disulfide bonds in LINC complexes play important roles in force transmission across the nuclear envelope. Other properties of LINC complexes, such as the helices preceding the SUN domain, the longer coiled-coils spanning the perinuclear space and higher-order organization may also function to transmit mechanical forces generated by the cytoskeleton across the nuclear envelope.
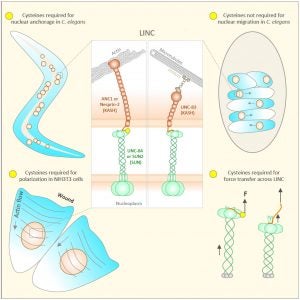
NE Cain, Z Jahed, A Schoenhofen, VA Valdez, B Elkin, H Hao, NJ Harris, LA Herrera, BM Woolums, MRK Mofrad, GWG Luxton, DA Starr (2018), Conserved SUN-KASH interactions mediate LINC complex-dependent nuclear movement and positioning. Current Biology.
Many nuclear positioning events involve linker of nucleoskeleton and cytoskeleton (LINC) complexes, which transmit forces generated by the cytoskeleton across the nuclear envelope. LINC complexes are formed by trans-luminal interactions between inner nuclear membrane SUN proteins and outer nuclear membrane KASH proteins, but how these interactions are regulated is poorly understood. We combine in vivo C. elegans genetics, in vitro wounded fibroblast polarization, and in silico molecular dynamic simulations to elucidate mechanisms of LINC complexes. The extension of the KASH domain by a single alanine residue or the mutation of the conserved tyrosine at -7 completely blocked the nuclear migration function of C. elegans UNC-83. Analogous mutations at -7 of mouse nesprin-2 disrupted rearward nuclear movements in NIH3T3 cells, but did not disrupt ANC-1 in nuclear anchorage. Furthermore, conserved cysteines predicted to form a disulfide bond between SUN and KASH proteins are important for the function of certain LINC complexes and might promote a developmental switch between nuclear migration and nuclear anchorage. Mutations of conserved cysteines in SUN or KASH disrupted ANC-1 dependent nuclear anchorage in C. elegans and Nesprin-2G dependent nuclear movements in polarizing fibroblasts. However, the SUN cysteine mutation did not disrupt nuclear migration. Moreover, molecular dynamic simulations showed that a disulfide bond is necessary for the maximal transmission of cytoskeleton-generated forces by LINC complexes in silico. Thus, we have demonstrated functions for SUN-KASH binding interfaces, including a predicted intermolecular disulfide bond, as mechanistic determinants of nuclear positioning and may represent targets for regulation.
HN Fridolfsson, LA Herrera, JN Brandt, NE Cain, GJ Herman, DA Starr (2018), Genetic analysis of nuclear migration and anchorage to study LINC complexes during development of Caenorhabditis elegans. G. Gundersen and H. Worman (eds) The LINC Complex. Methods in Molecular Biology.
Unlike the classical nuclear envelope with two membranes found in other eukaryotic cells, most nematode sperm nuclei are not encapsulated by membranes. Instead, they are surrounded by a nuclear halo of unknown composition. How the halo is formed and regulated is unknown. We used forward genetics to identify molecular lesions behind three classical fer (fertilization defective) mutations that disrupt the ultrastructure of the Caenorhabditis elegans sperm nuclear halo. We found fer-2 and fer-4 alleles to be nonsense mutations in mib-1. fer-3 was caused by a nonsense mutation in eri-3. GFP::MIB-1 was expressed in the germline during early spermatogenesis, but not in mature sperm. mib-1 encodes a conserved E3 ubiquitin ligase homologous to vertebrate Mib1 and Mib2, which function in Notch signaling. Here, we show that mib-1 is important for male sterility and is involved in the regulation or formation of the nuclear halo during nematode spermatogenesis.
LA Herrera, DA Starr (2018), The E3 ubiquitin ligase MIB-1 is necessary to form the nuclear halo in Caeorhabditis elegans sperm. G3: Genes/Genomes/Genetics.
Unlike the classical nuclear envelope with two membranes found in other eukaryotic cells, most nematode sperm nuclei are not encapsulated by membranes. Instead, they are surrounded by a nuclear halo of unknown composition. How the halo is formed and regulated is unknown. We used forward genetics to identify molecular lesions behind three classical fer (fertilization defective) mutations that disrupt the ultrastructure of the Caenorhabditis elegans sperm nuclear halo. We found fer-2 and fer-4 alleles to be nonsense mutations in mib-1. fer-3 was caused by a nonsense mutation in eri-3. GFP::MIB-1 was expressed in the germline during early spermatogenesis, but not in mature sperm. mib-1 encodes a conserved E3 ubiquitin ligase homologous to vertebrate Mib1 and Mib2, which function in Notch signaling. Here, we show that mib-1 is important for male sterility and is involved in the regulation or formation of the nuclear halo during nematode spermatogenesis.
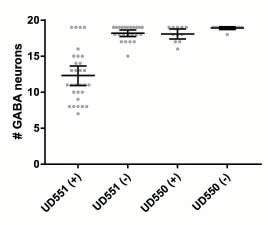
J Ho, VA Valdez, L Ma, DA Starr (2018), Characterizing Dynein’s role in P-cell nuclear migration using an auxin-induced degradation system. microPublication Biology.
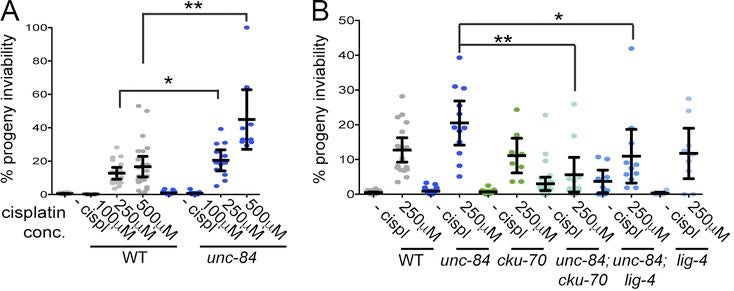
K Lawrence, EC Tapley, VE Cruz, Q Li, K Aung, KC Hart, TU Schwartz, DA Starr, J Engebrecht (2016), LINC complexes license homologous recombination in part through microtubule-dependent inhibition of nonhomologous end joining. The Journal of Cell Biology.
The Caenorhabditis elegans SUN domain protein, UNC-84, functions in nuclear migration and anchorage in the soma. We discovered a novel role for UNC-84 in DNA damage repair and meiotic recombination. Loss of UNC-84 leads to defects in the loading and disassembly of the recombinase RAD-51. Similar to mutations in Fanconi anemia (FA) genes, unc-84 mutants and human cells depleted of Sun-1 are sensitive to DNA cross-linking agents, and sensitivity is rescued by the inactivation of nonhomologous end joining (NHEJ). UNC-84 also recruits FA nuclease FAN-1 to the nucleoplasm, suggesting that UNC-84 both alters the extent of repair by NHEJ and promotes the processing of cross-links by FAN-1. UNC-84 interacts with the KASH protein ZYG-12 for DNA damage repair. Furthermore, the microtubule network and interaction with the nucleoskeleton are important for repair, suggesting that a functional linker of nucleoskeleton and cytoskeleton (LINC) complex is required. We propose that LINC complexes serve a conserved role in DNA repair through both the inhibition of NHEJ and the promotion of homologous recombination at sites of chromosomal breaks.
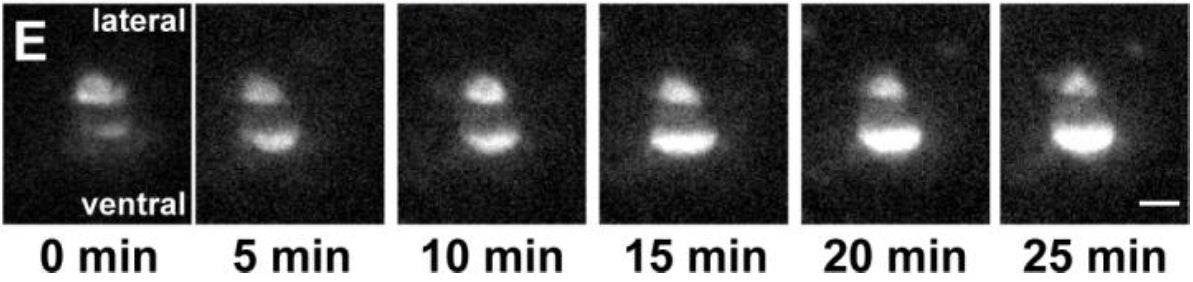
CR Bone, Y Chang, NE Cain, SP Murphy, DA Starr (2016), Nuclei migrate through constricted spaces using microtubule motors and actin networks in C. elegans hypodermal cells. Development.
Cellular migrations through constricted spaces are a critical aspect of many developmental and disease processes including hematopoiesis, inflammation, and metastasis. A limiting factor in these events is nuclear deformation. Here, we establish an in vivo model where nuclei can be visualized while moving through constrictions and use it to elucidate mechanisms for nuclear migration. C. elegans hypodermal P-cell larval nuclei traverse a narrow space about 5% their width. This constriction is blocked by fibrous organelles, structures connecting the muscles to cuticle through P cells. Fibrous organelles are removed just prior to nuclear migration, when nuclei and lamins undergo extreme morphological changes to squeeze through the space. Both actin and microtubule networks are organized to mediate nuclear migration. The LINC complex, consisting of the SUN protein UNC-84 and the KASH protein UNC-83, recruits dynein and kinesin-1 to the nuclear surface. Both motors function in P-cell nuclear migration, but dynein, functioning through UNC-83, plays a more central role as nuclei migrate toward minus ends of polarized microtubule networks. Thus, the nucleoskeleton and cytoskeleton are coordinated to move nuclei through constricted spaces.
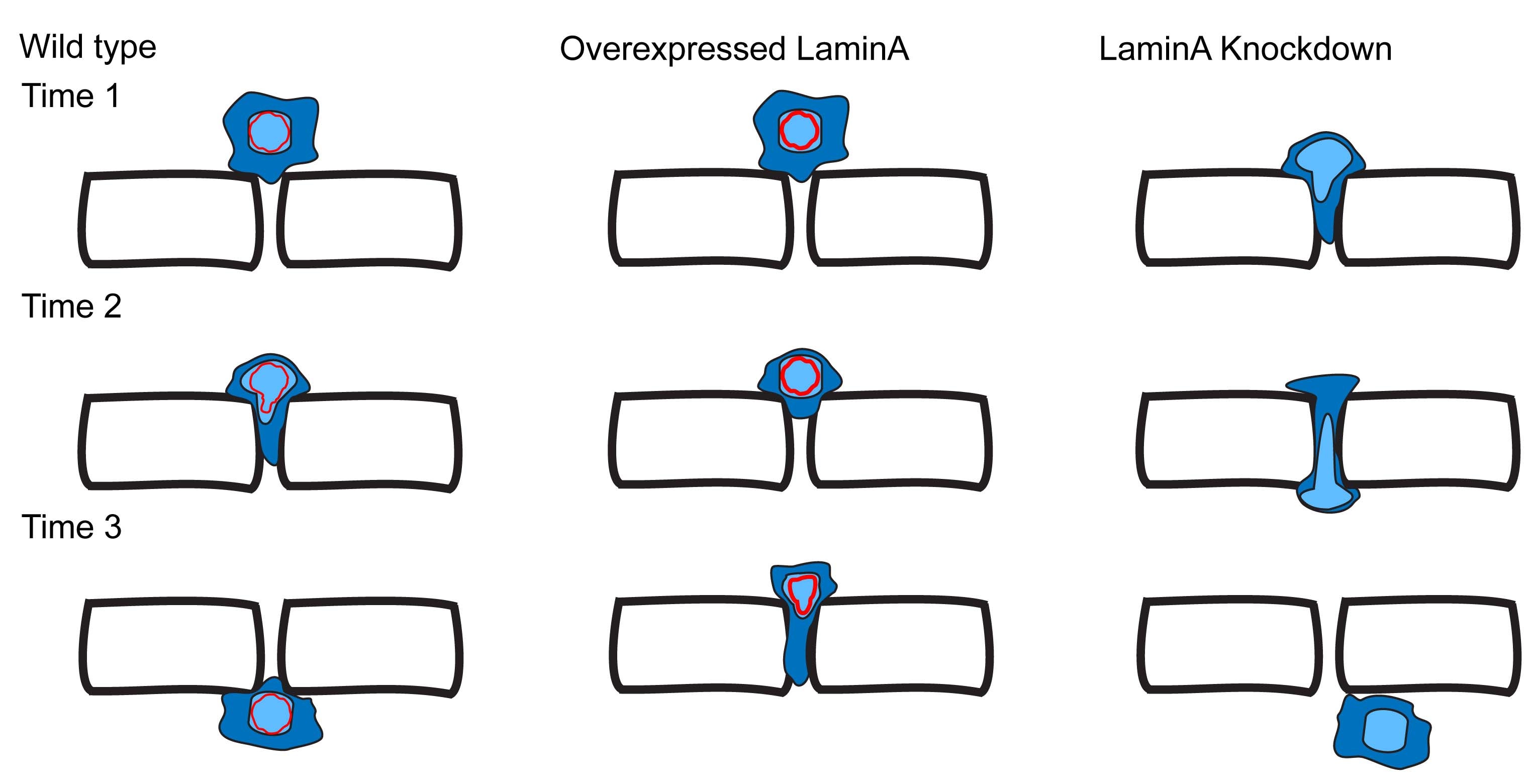
CR Bone, DA Starr (2016), Nuclear migration events throughout development. Journal of Cell Science.
Moving the nucleus to a specific position within the cell is an important event during many cell and developmental processes. Several different molecular mechanisms exist to position nuclei in various cell types. In this Commentary, we review the recent progress made in elucidating mechanisms of nuclear migration in a variety of important developmental models. Genetic approaches to identify mutations that disrupt nuclear migration in yeast, filamentous fungi, Caenorhabditis elegans, Drosophila and plants led to the identification of microtubule motors, as well as SUN and KASH proteins (LINC complex) that function to connect nuclei to the cytoskeleton. We focus on how these proteins and other mechanisms move nuclei throughout vertebrate development, including processes related to wound healing of fibroblasts, fertilization, developing myotubes and the developing central nervous system. We also describe how nuclear migration is involved in cells that migrate through constricted spaces. Based on these findings, it is becoming increasingly clear that defects in nuclear positioning are associated with human diseases.
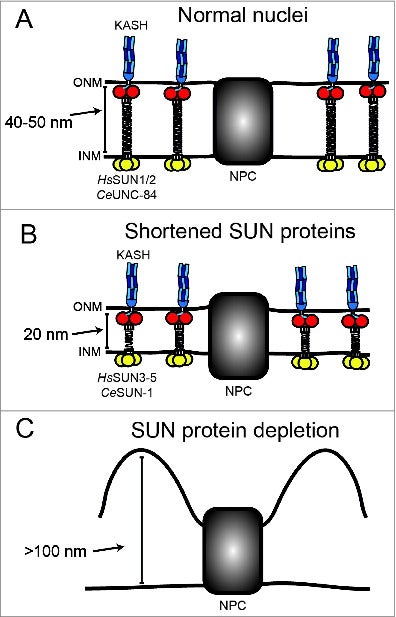
NE Cain, DA Starr (2014), SUN proteins and nuclear envelope spacing. Nucleus.
The nuclear envelope consists of 2 membranes separated by 30–50 nm, but how the 2 membranes are evenly spaced has been an open question in the field. Nuclear envelope bridges composed of inner nuclear membrane SUN proteins and outer nuclear membrane KASH proteins have been proposed to set and regulate nuclear envelope spacing. We tested this hypothesis directly by examining nuclear envelope spacing in Caenorhabditis elegans animals lacking UNC-84, the sole somatic SUN protein. SUN/KASH bridges are not required to maintain even nuclear envelope spacing in most tissues. However, UNC-84 is required for even spacing in body wall muscle nuclei. Shortening UNC-84 by 300 amino acids did not narrow the nuclear envelope space. While SUN proteins may play a role in maintaining nuclear envelope spacing in cells experiencing forces, our data suggest they are dispensable in most cells.
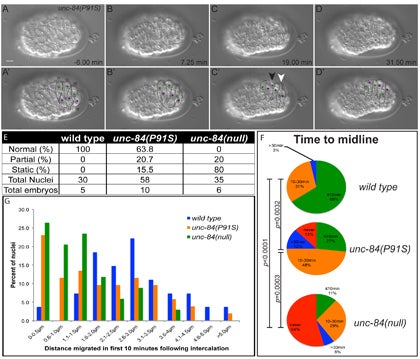
CR Bone, EC Tapley, M Gorjánácz, DA Starr (2014), The C. elegans SUN protein UNC-84 interacts with lamin to transfer forces from the cytoplasm to the nucleoskeleton during nuclear migration. Molecular Biology of the Cell.
Nuclear migration is a critical component of many cellular and developmental processes. The nuclear envelope forms a barrier between the cytoplasm, where mechanical forces are generated, and the nucleoskeleton. The LINC complex consists of KASH proteins in the outer nuclear membrane and SUN proteins in the inner nuclear membrane that bridge the nuclear envelope. How forces are transferred from the LINC complex to the nucleoskeleton is poorly understood. The C. elegans lamin, LMN-1, is required for nuclear migration and interacts with the nucleoplasmic domain of the SUN protein UNC-84. This interaction is weakened by the unc-84(P91S) missense mutation. These mutant nuclei have an intermediate nuclear migration defect—live imaging of nuclei or LMN-1::GFP show that many nuclei migrate normally, others initiate migration before subsequently failing, and others fail to begin migration. At least one other component of the nucleoskeleton, the NET5/Samp1/Ima1 homolog SAMP-1, plays a role in nuclear migration. We propose a nut and bolt model to explain how forces are dissipated across the nuclear envelope during nuclear migration. In this model, SUN/KASH bridges serve as bolts through the nuclear envelope, and nucleoskeleton components LMN-1 and SAMP-1 act as both nuts and washers on the inside of the nucleus.
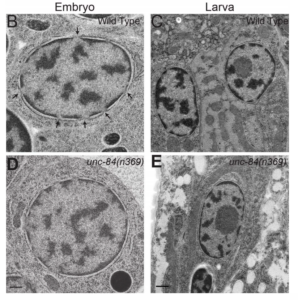
NE Cain, EC Tapley, KL McDonald, BM Cain, DA Starr (2014), The SUN protein UNC-84 is required only in force-bearing cells to maintain nuclear envelope architecture. The Journal of Cell Biology.
The nuclear envelope (NE) consists of two evenly spaced bilayers, the inner and outer nuclear membranes. The Sad1p and UNC-84 (SUN) proteins and Klarsicht, ANC-1, and Syne homology (KASH) proteins that interact to form LINC (linker of nucleoskeleton and cytoskeleton) complexes connecting the nucleoskeleton to the cytoskeleton have been implicated in maintaining NE spacing. Surprisingly, the NE morphology of most Caenorhabditis elegans nuclei was normal in the absence of functional SUN proteins. Distortions of the perinuclear space observed in unc-84 mutant muscle nuclei resembled those previously observed in HeLa cells, suggesting that SUN proteins are required to maintain NE architecture in cells under high mechanical strain. The UNC-84 protein with large deletions in its luminal domain was able to form functional NE bridges but had no observable effect on NE architecture. Therefore, SUN-KASH bridges are only required to maintain NE spacing in cells subjected to increased mechanical forces. Furthermore, SUN proteins do not dictate the width of the NE.
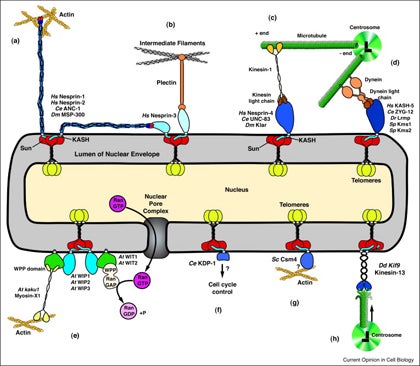
GWG Luxton, DA Starr (2014), KASHing up with the nucleus: novel functional roles of KASH proteins at the cytoplasmic surface of the nucleus. Current Opinion in Cell Biology.
Nuclear-cytoskeletal connections are central to fundamental cellular processes, including nuclear positioning and chromosome movements in meiosis. The cytoskeleton is coupled to the nucleoskeleton through conserved KASH-SUN bridges, or LINC complexes, that span the nuclear envelope. KASH proteins localize to the outer nuclear membrane where they connect the nucleus to the cytoskeleton. New findings have expanded the functional diversity of KASH proteins, showing that they interact with microtubule motors, actin, intermediate filaments, a nonconventional myosin, RanGAP, and each other. The role of KASH proteins in cellular mechanics is discussed. Genetic mutations in KASH proteins are associated with autism, hearing loss, cancer, muscular dystrophy and other diseases.
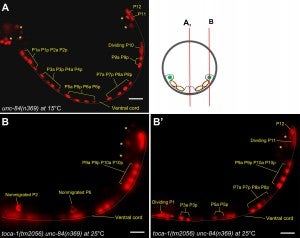
Y Chang, D Dranow, J Kuhn, M Meyerzon, M Ngo, D Ratner, K Warltier, DA Starr (2013), toca-1 is in a novel pathway that functions in parallel with a SUN-KASH nuclear envelope bridge to move nuclei in Caenorhabditis elegans. Genetics.
Moving the nucleus to an intracellular location is critical to many fundamental cell and developmental processes, including cell migration, differentiation, fertilization, and establishment of cellular polarity. Bridges of SUN and KASH proteins span the nuclear envelope and mediate many nuclear positioning events, but other pathways function independently through poorly characterized mechanisms. To identify and characterize novel mechanisms of nuclear migration, we conducted a non-biased forward genetic screen for mutations that enhanced the nuclear migration defect of unc-84, which encodes a SUN protein. In Caenorhabditis elegans larvae, failure of hypodermal P-cell nuclear migration results in uncoordinated and egg-laying defective animals. The process of P-cell nuclear migration in unc-84 null animals is temperature sensitive; at 25°C migration fails in unc-84mutants, but at 15°C the migration occurs normally. We hypothesized that an additional pathway functions in parallel to the unc-84 pathway to move P-cell nuclei at 15°C. In support of our hypothesis, forward genetic screens isolated eight emu (for enhancer of the nuclear migration defect of unc-84) mutations that only disrupt nuclear migration in a null unc-84 background. The yc20 mutant was determined to carry a mutation in the toca-1 gene. TOCA-1 functions to move P-cell nuclei in a cell-autonomous manner. TOCA-1 is conserved in humans, where it functions to nucleate and organize actin during endocytosis. Therefore, we have uncovered a player in a previously unknown, likely actin-dependent, pathway that functions to move nuclei in parallel to SUN-KASH bridges. The other emu mutations potentially represent other components of this novel pathway.
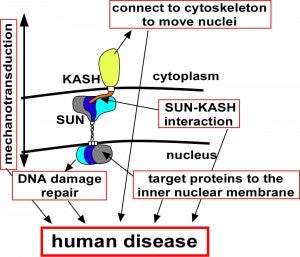
EC Tapley, DA Starr (2012), Connecting the nucleus to the cytoskeleton by SUN–KASH bridges across the nuclear envelope. Current Opinion in Cell Biology.
The nuclear–cytoskeleton connection influences many aspects of cellular architecture, including nuclear positioning, the stiffness of the global cytoskeleton, and mechanotransduction. Central to all of these processes is the assembly and function of conserved SUN–KASH bridges, or LINC complexes, that span the nuclear envelope. Recent studies provide details of the higher order assembly and targeting of SUN proteins to the inner nuclear membrane. Structural studies characterize SUN–KASH interactions that form the central link of the nuclear-envelope bridge. KASH proteins at the outer nuclear membrane link the nuclear envelope to the cytoskeleton where forces are generated to move nuclei. Significantly, SUN proteins were recently shown to contribute to the progression of laminopathies.
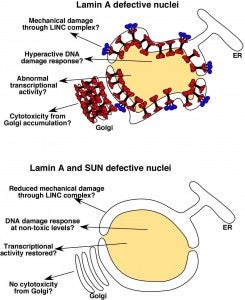
DA Starr (2012), Laminopathies: too much SUN is a bad thing. Current Biology.
SUN proteins accelerate the pathological progression of laminopathies. Although the mechanisms of how this occurs remain to be elucidated, an intriguing possibility is that high levels of Sun proteins lead to a hyperactive DNA damage response.
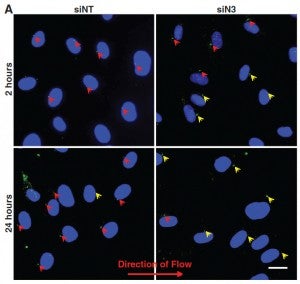
JT Morgan, ER Pfeiffer, TL Thirkill, G Peng, HN Fridolfsson, GC Douglas, DA Starr, AI Barakat (2011), Nesprin-3 regulates endothelial cell morphology, perinuclear cytoskeletal architecture, and flow-induced polarization. Molecular Biology of the Cell.
Changes in blood flow regulate gene expression and protein synthesis in vascular endothelial cells, and this regulation is involved in the development of atherosclerosis. How mechanical stimuli are transmitted from the endothelial luminal surface to the nucleus is incompletely understood. The linker of nucleus and cytoskeleton (LINC) complexes have been proposed as part of a continuous physical link between the plasma membrane and subnuclear structures. LINC proteins nesprin-1, -2, and -4 have been shown to mediate nuclear positioning via microtubule motors and actin. Although nesprin-3 connects intermediate filaments to the nucleus, no functional consequences of nesprin-3 mutations on cellular processes have been described. Here we show that nesprin-3 is robustly expressed in human aortic endothelial cells (HAECs) and localizes to the nuclear envelope. Nesprin-3 regulates HAEC morphology, with nesprin-3 knockdown inducing prominent cellular elongation. Nesprin-3 also organizes perinuclear cytoskeletal organization and is required to attach the centrosome to the nuclear envelope. Finally, nesprin-3 is required for flow-induced polarization of the centrosome and flow-induced migration in HAECs. These results represent the most complete description to date of nesprin-3 function and suggest that nesprin-3 regulates vascular endothelial cell shape, perinuclear cytoskeletal architecture, and important aspects of flow-mediated mechanotransduction.
EC Tapley, DA Starr (2011), Multiple mechanisms actively target the SUN protein UNC-84 to the inner nuclear membrane. Molecular Biology of the Cell.
Approximately 100 proteins are targeted to the inner nuclear membrane where they regulate chromatin and nuclear dynamics. However, the mechanisms underlying trafficking to the inner nuclear membrane are poorly understood. The Caenorhabditis elegans SUN protein UNC-84 is an excellent model to investigate such mechanisms. UNC-84 recruits KASH proteins to the outer nuclear membrane to bridge the nuclear envelope, mediating nuclear positioning. UNC-84 has four targeting sequences: two classical nuclear localization signals, an inner nuclear membrane sorting motif, and a signal conserved in mammalian Sun1, the SUN-nuclear-envelope-localization signal. Mutations in some signals disrupt the timing of UNC-84 nuclear-envelope localization, showing that diffusion is not sufficient to move all UNC-84 to the nuclear envelope. Thus, targeting UNC-84 requires an initial step that actively transports UNC-84 from the peripheral ER to the nuclear envelope. Only when all four signals are simultaneously disrupted does UNC-84 completely fail to localize and to function in nuclear migration, meaning that at least three signals function, in part, redundantly to ensure proper targeting of UNC-84. Multiple mechanisms might also be used to target other proteins to the inner nuclear membrane, thereby ensuring their proper and timely localization for essential cellular and developmental functions.
R Green, HL Kao, A Audhya, S Arur, JR Mayers, H Fridolfsson, M Schulman, S Schloissnig, S Niessen, K Laband, S Wang, DA Starr, A Hyman, T Schedl, A Desai, F Piano, KC Gunsalus, K Oegema (2011), A high-resolution C. elegans essential gene network based on phenotypic profiling of a complex tissue. Cell.
High-content screening for gene profiling has generally been limited to single cells. Here, we explore an alternative approach-profiling gene function by analyzing effects of gene knockdowns on the architecture of a complex tissue in a multicellular organism. We profile 554 essential C. elegans genes by imaging gonad architecture and scoring 94 phenotypic features. To generate a reference for evaluating methods for network construction, genes were manually partitioned into 102 phenotypic classes, predicting functions for uncharacterized genes across diverse cellular processes. Using this classification as a benchmark, we developed a robust computational method for constructing gene networks from high-content profiles based on a network context-dependent measure that ranks the significance of links between genes. Our analysis reveals that multi-parametric profiling in a complex tissue yields functional maps with a resolution similar to genetic interaction-based profiling in unicellular eukaryotes-pinpointing subunits of macromolecular complexes and components functioning in common cellular processes.
HN Fridolfsson, DA Starr (2010), Kinesin-1 and dynein at the nuclear envelope mediate the bidirectional migrations of nuclei. The Journal of Cell Biology.
Check out our perspective in BioArchitecture
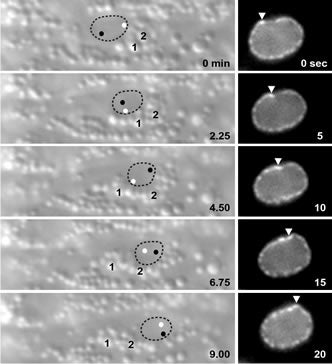
Kinesin-1 and dynein are recruited to the nuclear envelope by the Caenorhabditis elegans klarsicht/ ANC -1/Syne homology (KASH) protein UNC-83 to move nuclei. The mechanisms of how these motors are coordinated to mediate nuclear migration are unknown. Time-lapse differential interference contrast and fluorescence imaging of embryonic hypodermal nuclear migration events were used to characterize the kinetics of nuclear migration and determine microtubule dynamics and polarity. Wild-type nuclei display bidirectional movements during migration and are also able to roll past cytoplasmic granules. unc-83, unc-84, and kinesin-1 mutants have severe nuclear migration defects. Without dynein, nuclear migration initiates normally but lacks bidirectional movement and shows defects in nuclear rolling, implicating dynein in resolution of cytoplasmic roadblocks. Microtubules are highly dynamic during nuclear migration. EB1::green fluorescence protein imaging demonstrates that microtubules are polarized in the direction of nuclear migration. This organization of microtubules fits with our model that kinesin-1 moves nuclei forward and dynein functions to move nuclei backward for short stretches to bypass cellular roadblocks.
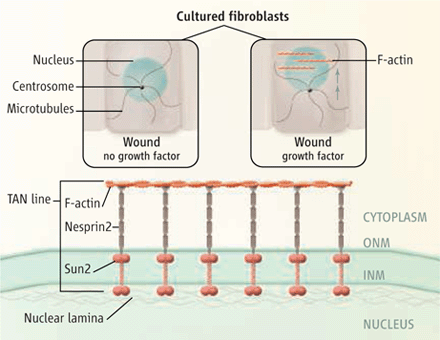
DA Starr (2010), Nuclei get TAN lines. Science invited perspective.
Positioning the nucleus within a cell involves a protein complex that spans the nuclear envelope to connect actin filaments to the nuclear lamina.
A perspective of Luxton et al. 2010. “Linear arrays of nuclear envelope proteins harness retrograde actin flow for nuclear movement.” Science 20; 956-959
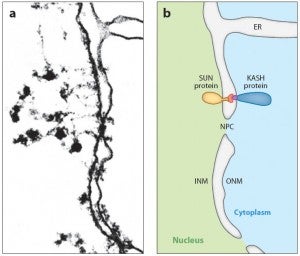
DA Starr, HN Fridolfsson (2010), Interactions between Nuclei and the Cytoskeleton are Mediated by SUN-KASH Nuclear-Envelope Bridges. Annual Review of Cell and Developmental Biology.
The nuclear envelope links the cytoskeleton to structural components of the nucleus. It functions to coordinate nuclear migration and anchorage, organize chromatin, and aid meiotic chromosome pairing. Forces generated by the cytoskeleton are transferred across the nuclear envelope to the nuclear lamina through a nuclear-envelope bridge consisting of SUN (Sad1 and UNC-84) and KASH proteins (Klarsicht, ANC-1 and Syne/Nesprin homology). Some KASH-SUN combinations connect microtubules, centrosomes, actin filaments, or intermediate filaments to the surface of the nucleus. Other combinations are used in cell cycle control, nuclear import, or apoptosis. Interactions between the cytoskeleton and the nucleus also affect global cytoskeleton organization. SUN and KASH proteins were identified through genetic screens for mispositioned nuclei in model organisms. Knockouts of SUN or KASH proteins disrupt neurological and muscular development in mice. Defects in SUN and KASH proteins have been linked to human diseases including muscular dystrophy, ataxia, progeria, lissencephaly, and cancer.
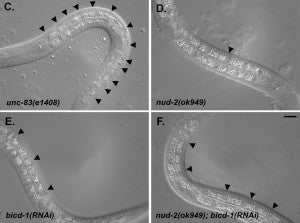
HN Fridolfsson, N Ly, M Meyerzon, DA Starr (2010), UNC-83 coordinates kinesin-1 and dynein activities at the nuclear envelope during nuclear migration. Developmental Biology.
Nuclei migrate during many events, including fertilization, establishment of polarity, differentiation, and cell division. The C. elegans KASH protein UNC-83 localizes to the outer nuclear membrane where it recruits kinesin-1 to provide the major motor activity required for nuclear migration in embryonic hyp7 cells. Here we show that UNC-83 also recruits two dynein-regulating complexes to the cytoplasmic face of the nucleus that play a regulatory role. One consists of the NudE homolog NUD-2 and the NudF/Lis1/Pac1 homolog LIS-1; the other includes dynein light chain DLC-1, the BicaudalD homolog BICD-1, and the egalitarian homologue EGAL-1. Genetic disruption of any member of these two complexes caused nuclear migration defects that were enhanced in some double mutant animals, suggesting that BICD-1 and EGAL-1 function in parallel to NUD-2. Dynein heavy chain mutant animals also had a nuclear migration defect, suggesting these complexes function through dynein. Deletion analysis indicated that independent domains of UNC-83 interact with kinesin and dynein. These data suggest a model where UNC-83 acts as the cargo-specific adaptor between the outer nuclear membrane and the microtubule motors kinesin-1 and dynein. Kinesin-1 functions as the major force generator during nuclear migration, while dynein is involved in regulation of bidirectional transport of the nucleus.
MD McGee, I Stagljar, DA Starr (2009), KDP-1 is a nuclear envelope KASH protein required for cell cycle progression. Journal of Cell Science.
KASH proteins localize to the outer nuclear membrane where they connect the nucleus to the cytoskeleton. KASH proteins interact with SUN proteins to transfer forces across the nuclear envelope to position nuclei or move chromosomes. A new KASH protein, KDP-1, was identified in a membrane yeast two-hybrid screen of a C. elegans library using the SUN protein UNC-84 as bait. KDP-1 also interacted with SUN-1. KDP-1 was enriched at the nuclear envelope in a variety of tissues and required SUN-1 for nuclear envelope localization in the germline. Genetic analyses showed that kdp-1 was essential for embryonic viability, larval growth, and germline development. kdp-1(RNAi) delayed the entry into mitosis in embryos, led to a small mitotic zone in the germline, and caused an endomitotic phenotype. Aspects of these phenotypes were similar to those seen in sun-1(RNAi) , suggesting that KDP-1 functions with SUN-1 in the germline and early embryo. The data suggest that KDP-1 is a novel KASH protein that functions to ensure the timely progression of the cell cycle between the end of S phase and the entry into mitosis.
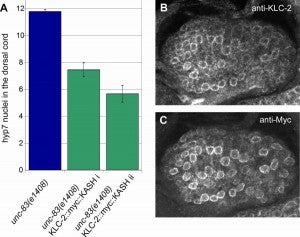
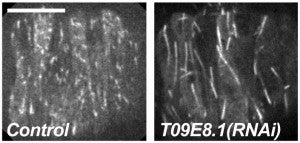
M Meyerzon, HN Fridolfsson, N Ly, FJ McNally, DA Starr (2009), UNC-83 is a nuclear-specific cargo adaptor for kinesin-1 mediated nuclear migration. Development.
Intracellular nuclear migration is essential for many cellular events including fertilization, establishment of polarity, division, and differentiation. How nuclei migrate is not understood at the molecular level. The C. elegans KASH protein UNC-83 is required for nuclear migration and localizes to the outer nuclear membrane. UNC-83 interacts with the inner nuclear membrane SUN protein, UNC-84, and is proposed to connect the cytoskeleton to the nuclear lamina. Here, we show that UNC-83 also interacts with the kinesin-1 light chain KLC-2 as identified in a yeast two-hybrid screen and confirmed by in vitro assays. UNC-83 interacts with and recruits KLC-2 to the nuclear envelope in a heterologous tissue culture system. Additionally, analysis of mutant phenotypes demonstrated that both KLC-2 and the kinesin-1 heavy chain, UNC-116, are required for nuclear migration. Finally, we can partially bypass the requirement for UNC-83 in nuclear migration by expressing a synthetic outer nuclear membrane KLC-2::KASH fusion protein. Our data support a model where UNC-83 plays a central role in nuclear migration by acting to bridge the nuclear envelope and as a kinesin-1 cargo-specific adaptor so that motor-generated forces specifically move the nucleus as a single unit.
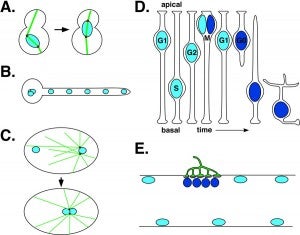
DA Starr (2009), A nuclear-envelope bridge positions nuclei and moves chromosomes. Journal of Cell Science.
Positioning the nucleus is essential for the formation of polarized cells, pronuclear migration, cell division, cell migration and the organization of specialized syncytia such as mammalian skeletal muscles. Proteins that are required for nuclear positioning also function during chromosome movements and pairing in meiosis. Defects in these processes lead to human diseases including laminopathies. To properly position the nucleus or move chromosomes within the nucleus, the cell must specify the outer surface of the nucleus and transfer forces across both membranes of the nuclear envelope. KASH proteins are specifically recruited to the outer nuclear membrane by SUN proteins, which reside in the inner nuclear membrane. KASH and SUN proteins physically interact in the perinuclear space, forming a bridge across the two membranes of the nuclear envelope. The divergent N-terminal domains of KASH proteins extend from the surface of the nucleus into the cytoplasm and interact with the cytoskeleton while the N termini of SUN proteins extend into the nucleoplasm to interact with the lamina or chromatin. The SUN and KASH bridge across the nuclear envelope functions to transfer forces generated in the cytoplasm into the nucleoplasm during nuclear migration, nuclear anchorage, centrosome attachment, intermediate-filament association and telomere clustering.
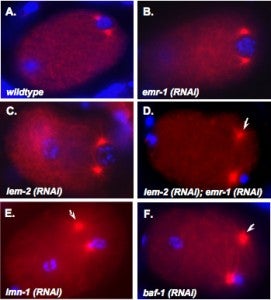
M Meyerzon, Z Gao, J Liu, J Wu, CJ Malone, DA Starr (2009), Centrosome attachment to the C. elegans male pronucleus is dependent on the surface area of the nuclear envelope. Developmental Biology.
A close association must be maintained between the male pronucleus and the centrosomes during pronuclear migration. In C. elegans, simultaneous depletion of inner nuclear membrane LEM proteins EMR-1 and LEM-2, depletion of the nuclear lamina proteins LMN-1 or BAF-1, or the depletion of nuclear import components leads to embryonic lethality with small pronuclei. Here, a novel centrosome detachment phenotype in C. elegans zygotes is described. Zygotes with defects in the nuclear envelope had small pronuclei with a single centrosome detached from the male pronucleus. ZYG-12, SUN-1, and LIS-1, which function at the nuclear envelope with dynein to attach centrosomes, were observed at normal concentrations on the nuclear envelope of pronuclei with detached centrosomes. Analysis of time-lapse images showed that as mutant pronuclei grew in surface area, they captured detached centrosomes. Larger tetraploid or smaller histone::mCherry pronuclei suppressed or enhanced the centrosome detachment phenotype respectively. In embryos fertilized with anucleated sperm, only one centrosome was captured by small female pronuclei, suggesting the mechanism of capture is dependent on the surface area of the outer nuclear membrane available to interact with aster microtubules. We propose that the limiting factor for centrosome attachment to the surface of abnormally small pronuclei is dynein.
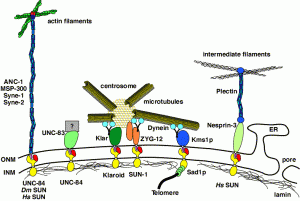
DA Starr (2007), Communication between the cytoskeleton and the nuclear envelope to position the nucleus. Molecular BioSystems.
In most eukaryotic cells, the nucleus is localized to a specific location. This highlight article focuses on recent advances describing the mechanisms of nuclear migration and anchorage. Central to nuclear positioning mechanisms is the communication between the nuclear envelope and the cytoskeleton. All three components of the cytoskeleton, microtubules, actin filaments and intermediate filaments, are involved in nuclear positioning to varying degrees in different cell types. KASH proteins on the outer nuclear membrane connect to SUN proteins on the inner nuclear membrane. Together they transfer forces between the cytoskeleton and the nuclear lamina. Once at the outer nuclear membrane, KASH proteins can interact with the cytoskeleton. Nuclear migrations are a component of many cellular migration events and defects in nuclear positioning lead to human diseases, most notably lissencephaly.
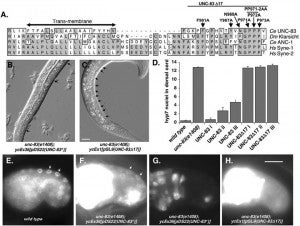
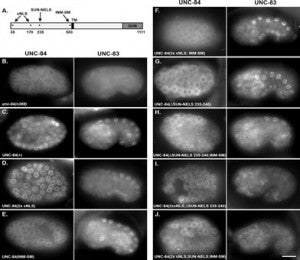
MD McGee*, R Rillo*, AS Anderson, DA Starr (2006), UNC-83 is a KASH protein required for nuclear migration and is recruited to the outer nuclear membrane by a physical interaction with the SUN protein UNC-84. Molecular Biology of the Cell. (*Equal contribution)
UNC-84 is required to localize UNC-83 to the nuclear envelope where it functions during nuclear migration. A KASH domain in UNC-83 was identified. KASH domains are conserved in the nuclear envelope proteins Syne/nesprins, Klarsicht, MSP-300, and ANC-1. C. elegans UNC-83 was shown to localize to the outer nuclear membrane and UNC-84 to the inner nuclear membrane in transfected mammalian cells, suggesting the KASH and SUN protein targeting mechanisms are conserved. Deletion of the KASH domain of UNC-83 blocked nuclear migration and localization to the C. elegans nuclear envelope. Some point mutations in the UNC-83 KASH domain disrupted nuclear migration, even if they localized normally. At least two separable portions of the C-terminal half of UNC-84 were found to interact with the UNC-83 KASH domain in a membrane-bound, split-ubiquitin yeast two-hybrid system. However, the SUN domain was essential for UNC-84 function and UNC-83 localization in vivo. These data support the model that KASH and SUN proteins bridge the nuclear envelope, connecting the nuclear lamina to cytoskeletal components. This mechanism appears conserved across eukaryotes and is the first proposed mechanism to target proteins specifically to the outer nuclear membrane.
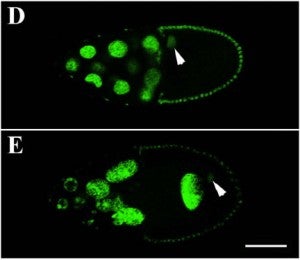
J Yu, DA Starr, X Wu, SM Parkhurst, Y Zhuang, T Xu, R Xu, M Han (2006), The KASH domain protein MSP-300 plays an essential role in nuclear anchoring during Drosophila oogenesis. Developmental Biology.
During late stages of Drosophila oogenesis, the cytoplasm of nurse cells in the egg chamber is rapidly transferred (‘‘dumped’’) to oocytes, while the nurse cell nuclei are anchored by a mechanism that involves the actin cytoskeleton. The factors that mediate this interaction between nuclei and actin cytoskeleton are unknown. MSP-300 is the likely Drosophila ortholog of the mammalian Syne-1 and -2 and C. elegans ANC-1 proteins, contained both actin-binding and nuclear envelope localization domains. By using an antibody against C-terminus of MSP-300, we find that MSP-300 is distributed throughout the cytoplasm and accumulates at the nuclear envelope of nurse cells and the oocyte. A GFP fusion protein containing the C-terminal region of MSP-300 is also sufficient to localize protein on the nuclear envelope in oocytes. To eliminate the maternal gene activity during oogenesis, we generated homozygous germ-line clones of a loss-of-function mutation in msp-300 in otherwise heterozygous mothers. In the mutant egg chambers that develop from such clones, cytoplasmic dumping of nurse cells is severely disturbed. The nuclei of nurse cells and the oocyte are mislocalized and the usually well-organized actin structures are severely disrupted. These results indicate that maternal MSP-300 plays an important role in actin-dependent nuclear anchorage during cytoplasmic transport.
DA Starr, JA Fischer (2005), KASH ‘n Karry: the KASH domain family of cargo-specific cytoskeletal adaptor proteins. Bioessays.
A diverse family of proteins has been discovered with a small C-terminal KASH domain in common. KASH domain proteins are localized uniquely to the outer nuclear envelope, enabling their cytoplasmic extensions to tether the nucleus to actin filaments or microtubules. KASH domains are targeted to the outer nuclear envelope by SUN domains of inner nuclear envelope proteins. Several KASH protein genes were discovered as mutant alleles in model organisms with defects in developmentally regulated nuclear positioning. Recently, KASH-less isoforms have been found that connect the cytoskeleton to organelles other than the nucleus. A widened view of these proteins is now emerging, where KASH proteins and their KASH-less counterparts are cargo-specific adaptors that not only link organelles to the cytoskeleton but also regulate developmentally specific organelle movements.
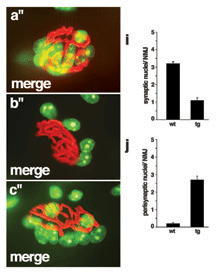
RM Grady*, DA Starr*, G Ackerman, JR Sanes, M Han (2005), Syne proteins anchor muscle nuclei at the neuromuscular junction. Proceedings of the National Academy of Sciences. (*Equal contribution)
MA Ruegg (2005), Organization of synaptic myonuclei by Syne proteins and their role during the formation of the nerve–muscle synapse. Commentary in Proceedings of the National Academy of Sciences.
Vertebrate skeletal muscle fibers contain hundreds of nuclei, of which three to six are functionally specialized and stably anchored beneath the postsynaptic membrane at the neuromuscular junction (NMJ). The mechanisms that localize synaptic nuclei and the roles they play in neuromuscular development are unknown. Syne-1 is concentrated at the nuclear envelope of synaptic nuclei; its Caenorhabditis elegans orthologue ANC-1 functions to tether nuclei to the cytoskeleton. To test the involvement of Syne proteins in nuclear anchoring, we generated transgenic mice overexpressing the conserved C-terminal Klarsicht ANC-1 Syne homology domain of Syne-1. The transgene acted in a dominant interfering fashion, displacing endogenous Syne-1 from the nuclear envelope. Muscle nuclei failed to aggregate at the NMJ in transgenic mice, demonstrating that localization and positioning of synaptic nuclei require Syne proteins. We then exploited this phenotype to show that synaptic nuclear aggregates are dispensable for maturation of the NMJ.
DA Starr, M Han (2005), A genetic approach to study the role of nuclear envelope components in nuclear positioning. Nuclear organization in development and disease, Wiley (Novartis Found Symp 264).
In many cell types, the nucleus is positioned to a specific location. Our work and others have demonstrated that several integral nuclear envelope proteins function to move the nucleus and to anchor it in place. Our forward genetic approach has identified three components of the nuclear envelope involved in nuclear positioning. ANC-1 consists of two actin-binding calponin domains, a huge central coiled domain, and a nuclear envelope targeting domain termed the KASH domain. ANC-1 functions to physically tether the actin cytoskeleton to the outer nuclear membrane. UNC-83 is a novel protein that functions in an unknown manner during nuclear migration. UNC-83 contains a domain with weak homology to the KASH domain of ANC-1. UNC-84 is a SUN protein that is required for both nuclear migration and anchorage. UNC-84 recruits both UNC-83 and ANC-1 to the nuclear envelope. We propose a model where UNC-84 is an integral component of the inner nuclear membrane, with its SUN domain in the perinuclear space. The SUN domain then recruits ANC-1 and UNC-83, through interactions with their KASH domains, to the outer nuclear envelope. Together these proteins function to bridge the two membranes of the nuclear envelope, connecting the nuclear matrix to the cytoskeleton.
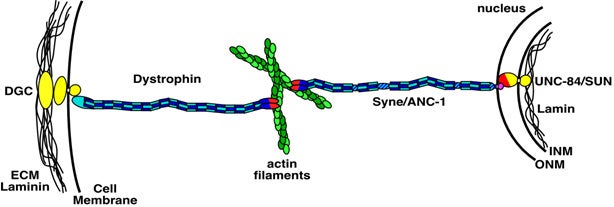
DA Starr, M Han (2003), ANChors away: an actin based mechanism of nuclear positioning. Journal of Cell Science.
Mechanisms for nuclear migration and nuclear anchorage function together to control nuclear positioning. Both tubulin and actin networks play important roles in nuclear positioning. The actin cytoskeleton has been shown to position nuclei in a variety of systems from yeast to plants and animals. It can either act as a stable skeleton to anchor nuclei or supply the active force to move nuclei. Two C. elegans genes and their homologues play important roles in these processes. Syne/ANC-1 anchors nuclei by directly tethering the nuclear envelope to the actin cytoskeleton, and UNC-84/SUN functions at the nuclear envelope to recruit Syne/ANC-1.
DA Starr, M Han (2002), Role of ANC-1 in tethering nuclei to the actin cytoskeleton. [Supplement] Science.
Mutations in anc-1 (nuclear anchorage defective) disrupt the positioning of nuclei and mitochondria in Caenorhabditis elegans. ANC-1 is shown to consist of mostly coiled regions with a nuclear envelope localization domain (called the KASH domain) and an actin-binding domain; this structure was conserved with the Drosophila protein Msp-300 and the mammalian Syne proteins. Antibodies against ANC-1 localized cytoplasmically and were enriched at the nuclear periphery in an UNC-84–dependent manner. Overexpression of the KASH domain or the actin-binding domain caused a dominant negative anchorage defect. Thus, ANC-1 may connect nuclei to the cytoskeleton by interacting with UNC-84 at the nuclear envelope and with actin in the cytoplasm.
KK Lee*, DA Starr*, M Cohen, J Liu, M Han, KL Wilson, Y Gruenbaum (2002), Lamin-dependent localization of UNC-84, a protein required for nuclear migration in C. elegans. Molecular Biology of the Cell. (*Equal contribution)
Mutations in the Caenorhabditis elegans unc-84 gene cause defects in nuclear migration and anchoring. We show that endogenous UNC-84 protein colocalizes with Ce-lamin at the nuclear envelope and that the envelope localization of UNC-84 requires Ce-lamin. We also show that during mitosis, UNC-84 remains at the nuclear periphery until late anaphase, similar to known inner nuclear membrane proteins. UNC-84 protein is first detected at the 26-cell stage and thereafter is present in most cells during development and in adults. UNC-84 is properly expressed in unc-83 and anc-1 lines, which have phenotypes similar to unc-84, suggesting that neither the expression nor nuclear envelope localization of UNC-84 depends on UNC-83 or ANC-1 proteins. The envelope localization of Ce-lamin, Ce-emerin, Ce-MAN1, and nucleoporins are unaffected by the loss of UNC-84. UNC-84 is not required for centrosome attachment to the nucleus because centrosomes are localized normally in unc-84 hyp7 cells despite a nuclear migration defect. Models for UNC-84 localization are discussed.
DS Fay, DA Starr, A Spencer, W Johnson, Edited by A Fluet, J Yochem (2001), Worm Breeding for Super Geniuses: A guide to genetic mapping in C. elegans.
What this guide is and isn’t. This guide is neither a basic text in Mendelian genetics nor is it in any way a comprehensive description of C. elegans biology. Detailed information about the latter can be gleaned from C. elegans I and C. elegans II by Cold Spring Harbor Press Inc. This guide does assume a working knowledge of basic genetics and will be of limited use to those who lack some background in this area. It is our hope that this text may serve as a supplement to existing published materials and that it will facilitate the successful breeding of worms by those new to the field.
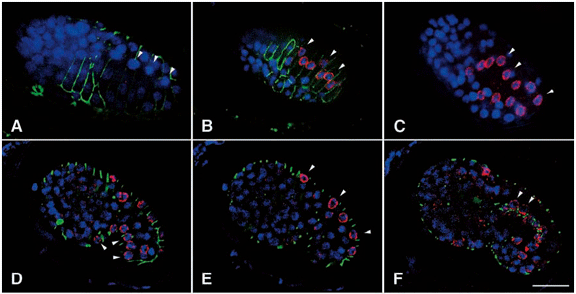
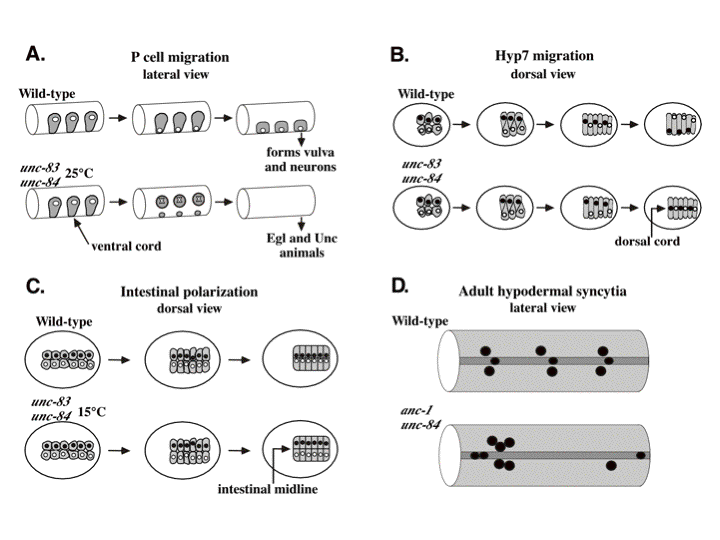
DA Starr, GJ Hermann, CJ Malone, W Fixsen, JR Priess, R Horvitz, M Han (2001), unc-83 encodes a novel component of the nuclear envelope and is essential for proper nuclear migration. Development.
Nuclear migration plays an essential role in the growth and development of a wide variety of eukaryotes. Mutations in unc-84, which encodes a conserved component of the nuclear envelope, have been shown to disrupt nuclear migration in two C. elegans tissues. We show that mutations in unc-83 disrupt nuclear migration in a similar manner in migrating P cells, hyp7 precursors and the intestinal primordium, but have no obvious defects in the association of centrosomes with nuclei or the structure of the nuclear lamina of migrating nuclei. We also show that unc-83 encodes a novel transmembrane protein. We identified three unc-83 transcripts that are expressed in a tissuespecific manner. Antibodies against UNC-83 co-localized to the nuclear envelope with lamin and UNC-84. Unlike UNC- 84, UNC-83 localized to only specific nuclei, many of which were migratory. UNC-83 failed to localize to the nuclear envelope in unc-84 mutants with lesions in the conserved SUN domain of UNC-84, and UNC-83 interacted with the SUN domain of UNC-84 in vitro, suggesting that these two proteins function together during nuclear migration. We favor a model in which UNC-84 directly recruits UNC-83 to the nuclear envelope where they help transfer force between the cytoskeleton and the nucleus.
Pre — 2021 Luxton Publications Google Scholar
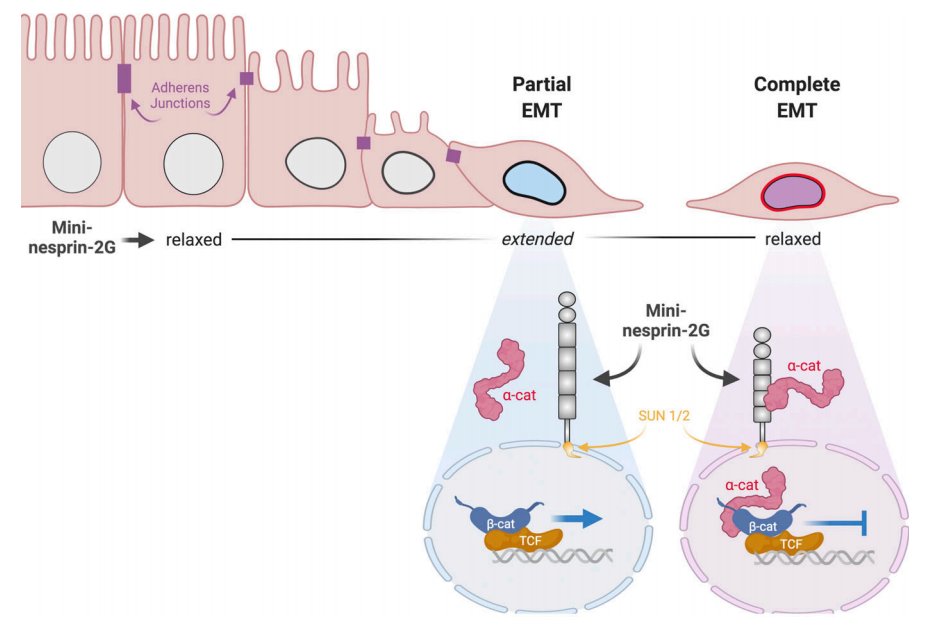
CJ Gottardi, GWG Luxton (2020), Nesprin-2G tension fine-tunes Wnt/β-catenin signaling. Journal of Cell Biology.
How LINC complexes mediate nuclear mechanotransduction remains unclear. In this issue, Déjardin, Carollo, et al. (2020. J. Cell Biol.) show that the LINC complex protein nesprin-2G is a mechanosensor of epithelial–mesenchymal transitions (EMTs), recruiting α-catenin to the nucleus to attenuate Wnt/β-catenin signaling.
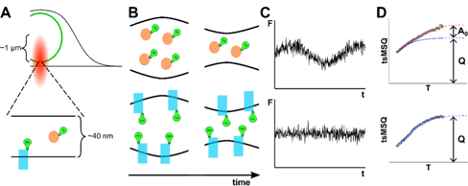
J Hennen, KH Hur, J Kohler, GWG Luxton, JD Mueller (2020), Differentiating Membrane-associated and Soluble Protein Populations within the Nuclear Envelope via Fluorescence Fluctuation Spectroscopy. Biophysical Journal.
The nuclear envelope (NE) consists of two membranes separated by the ∼40 nm wide lumen. This thin structure, being well below the resolution limit, presents a challenge to differentiating membrane-associated and luminal protein populations within the NE of living cells. Distinguishing these two populations is further complicated by the observation that diffusion time, a commonly used marker for membrane association in other cellular compartments, shows a strong dependence on molecular size within the lumen. Thus, a decrease in proteins mobility within the NE may be caused by the formation of large soluble complexes or association with the membranes. We recently observed a striking divergence in the temperature dependence of mobility for luminal and membrane-associated proteins within the NE, which provides a reliable marker for differentiating these two populations. Furthermore, previously reported nuclear membrane undulations introduce an additional fluctuation signal to luminal proteins while not affecting membrane-associated proteins. We demonstrate that this effect providing a second, independent marker distinguishing luminal and membrane-associated protein. These two independent assays for membrane-association are applied to proteins of biological interest. The luminal domain of SUN2 was previously shown to exist as a mixture of fast diffusing monomers and slowly diffusing trimers. We apply our tools to determine whether this decrease in mobility is caused solely by the three-fold increase in molecular weight or by membrane association. Finally, we use these techniques to identify the extent of membrane association for the AAA+ ATPase torsinA and explore the effect of tagging the fluorescent label to its N- or C-terminus. This work has been supported by a grant from the National Institutes of Health (R01 GM64589).
JE Hölper, BG Klupp, GWG Luxton, K Franzke, TC Mettenleiter (2020), Function of Torsin AAA+ ATPases in Pseudorabies Virus Nuclear Egress. Cells.
Newly assembled herpesvirus nucleocapsids traverse the intact nuclear envelope by a vesicle-mediated nucleo-cytoplasmic transport for final virion maturation in the cytoplasm. For this, they bud at the inner nuclear membrane resulting in primary enveloped particles in the perinuclear space (PNS) followed by fusion of the primary envelope with the outer nuclear membrane (ONM). While the conserved viral nuclear egress complex orchestrates the first steps, effectors of fusion of the primary virion envelope with the ONM are still mostly enigmatic but might include cellular proteins like SUN2 or ESCRT-III components. Here, we analyzed the influence of the only known AAA+ ATPases located in the endoplasmic reticulum and the PNS, the Torsins (Tor), on nuclear egress of the alphaherpesvirus pseudorabies virus. For this overexpression of wild type and mutant proteins as well as CRISPR/Cas9 genome editing was applied. Neither single overexpression nor gene knockout (KO) of TorA or TorB had a significant impact. However, TorA/B double KO cells showed decreased viral titers at early time points of infection and an accumulation of primary virions in the PNS pointing to a delay in capsid release during nuclear egress.
J Hennen, KH Hur, J Kohler, SR Karuka, I Angert, GWG Luxton, JD Mueller (2019), Identifying heteroprotein complexes in the nuclear envelope. Biophysical Journal.
The nucleus is delineated by the nuclear envelope (NE), which is a double membrane barrier composed of the inner and outer nuclear membranes as well as a ∼40-nm wide lumen. In addition to its barrier function, the NE acts as a critical signaling node for a variety of cellular processes, which are mediated by protein complexes within this subcellular compartment. Although fluorescence fluctuation spectroscopy is a powerful tool for characterizing protein complexes in living cells, it was recently demonstrated that conventional fluorescence fluctuation spectroscopy methods are not suitable for applications in the NE because of the presence of slow nuclear membrane undulations. We previously addressed this challenge by developing time-shifted mean-segmented Q (tsMSQ) analysis and applied it to successfully characterize protein homo-oligomerization in the NE. However, many NE complexes, such as the linker of the nucleoskeleton and cytoskeleton complex, are formed by heterotypic interactions, which single-color tsMSQ is unable to characterize. Here, we describe the development of dual-color (DC) tsMSQ to analyze NE heteroprotein complexes built from proteins that carry two spectrally distinct fluorescent labels. Experiments performed on model systems demonstrate that DC tsMSQ properly identifies heteroprotein complexes and their stoichiometry in the NE by accounting for spectral cross talk and local volume fluctuations. Finally, we applied DC tsMSQ to study the assembly of the linker of the nucleoskeleton and cytoskeleton complex, a heteroprotein complex composed of Klarsicht/ANC-1/SYNE homology and Sad1/UNC-84 (SUN) proteins, in the NE of living cells. Using DC tsMSQ, we demonstrate the ability of the SUN protein SUN2 and the Klarsicht/ANC-1/SYNE homology protein nesprin-2 to form a heterocomplex in vivo. Our results are consistent with previously published in vitro studies and demonstrate the utility of the DC tsMSQ technique for characterizing NE heteroprotein complexes.
AJR Booth, Z Yue, JK Eykelenboom, T Stiff, GWG Luxton, H Hochegger, TU Tanaka (2019), Contractile acto-myosin network on nuclear envelope remnants positions human chromosomes for mitosis. eLife.
To ensure proper segregation during mitosis, chromosomes must be efficiently captured by spindle microtubules and subsequently aligned on the mitotic spindle. The efficacy of chromosome interaction with the spindle can be influenced by how widely chromosomes are scattered in space. Here, we quantify chromosome-scattering volume (CSV) and find that it is reduced soon after nuclear envelope breakdown (NEBD) in human cells. The CSV reduction occurs primarily independently of microtubules and is therefore not an outcome of interactions between chromosomes and the spindle. We find that, prior to NEBD, an acto-myosin network is assembled in a LINC complex-dependent manner on the cytoplasmic surface of the nuclear envelope. This acto-myosin network remains on nuclear envelope remnants soon after NEBD, and its myosin-II-mediated contraction reduces CSV and facilitates timely chromosome congression and correct segregation. Thus, we find a novel mechanism that positions chromosomes in early mitosis to ensure efficient and correct chromosome–spindle interactions.
NK Gill, C Ly, PH Kim, CA Saunders, LG Fong, SG Young, GWG Luxton, AC Rowat (2019), DYT1 Dystonia Patient-Derived Fibroblasts Have Increased Deformability and Susceptibility to Damage by Mechanical Forces. Frontiers in Cellular and Developmental Biology.
DYT1 dystonia is a neurological movement disorder that is caused by a loss-of-function mutation in the DYT1/TOR1A gene, which encodes torsinA, a conserved luminal ATPases-associated with various cellular activities (AAA+) protein. TorsinA is required for the assembly of functional linker of nucleoskeleton and cytoskeleton (LINC) complexes, and consequently the mechanical integration of the nucleus and the cytoskeleton. Despite the potential implications of altered mechanobiology in dystonia pathogenesis, the role of torsinA in regulating cellular mechanical phenotype, or mechanotype, in DYT1 dystonia remains unknown. Here, we define the deformability of mouse fibroblasts lacking functional torsinA as well as human fibroblasts isolated from DYT1 dystonia patients. We find that the deletion of torsinA or the expression of torsinA containing the DYT1 dystonia-causing ΔE302/303 (ΔE) mutation results in more deformable cells. We observe a similar increased deformability of mouse fibroblasts that lack lamina-associated polypeptide 1 (LAP1), which interacts with and stimulates the ATPase activity of torsinA in vitro, as well as with the absence of the LINC complex proteins, Sad1/UNC-84 1 (SUN1) and SUN2, lamin A/C, or lamin B1. Consistent with these findings, we also determine that DYT1 dystonia patient-derived fibroblasts are more compliant than fibroblasts isolated from unafflicted individuals. DYT1 dystonia patient-derived fibroblasts also exhibit increased nuclear strain and decreased viability following mechanical stretch. Taken together, our results establish the foundation for future mechanistic studies of the role of cellular mechanotype and LINC-dependent nuclear-cytoskeletal coupling in regulating cell survival following exposure to mechanical stresses.
W Chang, Y Wang, GWG Luxton, C Östlund, HJ Worman, GG Gundersen (2019), Imbalanced nucleocytoskeletal connections create common polarity defects in progeria and physiological aging. Proceedings of the National Academy of Sciences.
Studies of the accelerated aging disorder Hutchinson–Gilford progeria syndrome (HGPS) can potentially reveal cellular defects associated with physiological aging. HGPS results from expression and abnormal nuclear envelope association of a farnesylated, truncated variant of prelamin A called “progerin.” We surveyed the diffusional mobilities of nuclear membrane proteins to identify proximal effects of progerin expression. The mobilities of three proteins—SUN2, nesprin-2G, and emerin—were reduced in fibroblasts from children with HGPS compared with those in normal fibroblasts. These proteins function together in nuclear movement and centrosome orientation in fibroblasts polarizing for migration. Both processes were impaired in fibroblasts from children with HGPS and in NIH 3T3 fibroblasts expressing progerin, but were restored by inhibiting protein farnesylation. Progerin affected both the coupling of the nucleus to actin cables and the oriented flow of the cables necessary for nuclear movement and centrosome orientation. Progerin overexpression increased levels of SUN1, which couples the nucleus to microtubules through nesprin-2G and dynein, and microtubule association with the nucleus. Reducing microtubule-nuclear connections through SUN1 depletion or dynein inhibition rescued the polarity defects. Nuclear movement and centrosome orientation were also defective in fibroblasts from normal individuals over 60 y, and both defects were rescued by reducing the increased level of SUN1 in these cells or inhibiting dynein. Our results identify imbalanced nuclear engagement of the cytoskeleton (microtubules: high; actin filaments: low) as the basis for intrinsic cell polarity defects in HGPS and physiological aging and suggest that rebalancing the connections can ameliorate the defects.

NE Cain, Z Jahed, A Schoenhofen, VA Valdez, B Elkin, H Hao, NJ Harris, LA Herrera, BM Woolums, MRK Mofrad, GWG Luxton, DA Starr (2018), Conserved SUN-KASH interactions mediate LINC complex-dependent nuclear movement and positioning. Current Biology.
Many nuclear positioning events involve linker of nucleoskeleton and cytoskeleton (LINC) complexes, which transmit forces generated by the cytoskeleton across the nuclear envelope. LINC complexes are formed by trans-luminal interactions between inner nuclear membrane SUN proteins and outer nuclear membrane KASH proteins, but how these interactions are regulated is poorly understood. We combine in vivo C. elegans genetics, in vitro wounded fibroblast polarization, and in silico molecular dynamic simulations to elucidate mechanisms of LINC complexes. The extension of the KASH domain by a single alanine residue or the mutation of the conserved tyrosine at -7 completely blocked the nuclear migration function of C. elegans UNC-83. Analogous mutations at -7 of mouse nesprin-2 disrupted rearward nuclear movements in NIH3T3 cells, but did not disrupt ANC-1 in nuclear anchorage. Furthermore, conserved cysteines predicted to form a disulfide bond between SUN and KASH proteins are important for the function of certain LINC complexes and might promote a developmental switch between nuclear migration and nuclear anchorage. Mutations of conserved cysteines in SUN or KASH disrupted ANC-1 dependent nuclear anchorage in C. elegans and Nesprin-2G dependent nuclear movements in polarizing fibroblasts. However, the SUN cysteine mutation did not disrupt nuclear migration. Moreover, molecular dynamic simulations showed that a disulfide bond is necessary for the maximal transmission of cytoskeleton-generated forces by LINC complexes in silico. Thus, we have demonstrated functions for SUN-KASH binding interfaces, including a predicted intermolecular disulfide bond, as mechanistic determinants of nuclear positioning and may represent targets for regulation.
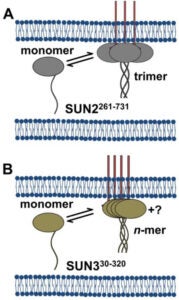
J Hennen, HK Ho, SR Karuka, LGW Gant, JD Mueller (2018), Protein oligomerization and mobility within the nuclear envelope evaluated by the time-shifted mean-segmented Q factor. Methods.
Analysis of fluorescence fluctuation experiments by the mean-segmented Q (MSQ) method was recently used to successfully characterize the oligomeric state and mobility of proteins within the nuclear envelope (NE) of living cells. However, two significant shortcomings of MSQ were recognized. Non-ideal detector behavior due to dead-time and afterpulsing as well as the lack of error analysis currently limit the potential of MSQ. This paper presents time-shifted MSQ (tsMSQ), a new formulation of MSQ that is robust with respect to dead-time and afterpulsing. In addition, a protocol for performing error analysis on tsMSQ data is introduced to assess the quality of fit models and estimate the uncertainties of fit parameters. Together, these developments significantly simplify and improve the analysis of fluorescence fluctuation data taken within the NE. To demonstrate these new developments, tsMSQ was used to characterize the oligomeric state and mobility of the luminal domains of two inner nuclear membrane SUN proteins. The results for the luminal domain of SUN2 obtained through tsMSQ without correction for non-ideal detector effects agree with a recent study that was conducted using the original MSQ formulation. Finally, tsMSQ was applied to characterize the oligomeric state and mobility of the luminal domain of the germline-restricted SUN3.
S Majumder, PT Willey, MS Denies, AP Liu*, GWG Luxton* (2018), A synthetic biology platform for the reconstitution and mechanistic dissection of LINC complex assembly. Journal of Cell Science. (*Equal contribution)
The linker of nucleoskeleton and cytoskeleton (LINC) is a conserved nuclear envelope-spanning molecular bridge that is responsible for the mechanical integration of the nucleus with the cytoskeleton. LINC complexes are formed by a transluminal interaction between the outer and inner nuclear membrane KASH and SUN proteins, respectively. Despite recent structural insights, our mechanistic understanding of LINC complex assembly remains limited by the lack of an experimental system for its in vitro reconstitution and manipulation. Here, we describe artificial nuclear membranes (ANMs) as a synthetic biology platform based on mammalian cell-free expression for the rapid reconstitution of SUN proteins in supported lipid bilayers. We demonstrate that SUN1 and SUN2 are oriented in ANMs with solvent-exposed C-terminal KASH-binding SUN domains. We also find that SUN2 possesses a single transmembrane domain, while SUN1 possesses three. Finally, SUN protein-containing ANMs bind synthetic KASH peptides, thereby reconstituting the LINC complex core. This work represents the first in vitro reconstitution of KASH-binding SUN proteins in supported lipid bilayers using cell-free expression, which will be invaluable for testing proposed models of LINC complex assembly and its regulation.
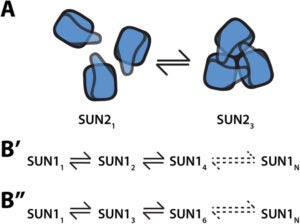
J Hennen, CA Saunders, JD Mueller*, GWG Luxton* (2018), Fluorescence fluctuation spectroscopy reveals differential SUN protein oligomerization in living cells. Molecular Biology of the Cell. (*Equal contributions)
Linker-of-nucleoskeleton-and-cytoskeleton (LINC) complexes are conserved molecular bridges within the nuclear envelope that mediate mechanical force transmission into the nucleoplasm. The core of a LINC complex is formed by a transluminal interaction between the outer and inner nuclear membrane KASH and SUN proteins, respectively. Mammals encode six KASH proteins and five SUN proteins. Recently, KASH proteins were shown to bind to the domain interfaces of trimeric SUN2 proteins in vitro. However, neither the existence of SUN2 trimers in living cells nor the extent to which other SUN proteins conform to this assembly state have been tested experimentally. Here we extend the application of fluorescence fluctuation spectroscopy to quantify SUN protein oligomerization in the nuclear envelopes of living cells. Using this approach, we demonstrate for the first time that SUN2 trimerizes in vivo and we demonstrate that the in vivo oligomerization of SUN1 is not limited to a trimer. In addition, we provide evidence to support the existence of potential regulators of SUN protein oligomerization in the nuclear envelope. The differential SUN protein oligomerization illustrated here suggests that SUN proteins may have evolved to form different assembly states in order to participate in diverse mechanotransduction events.
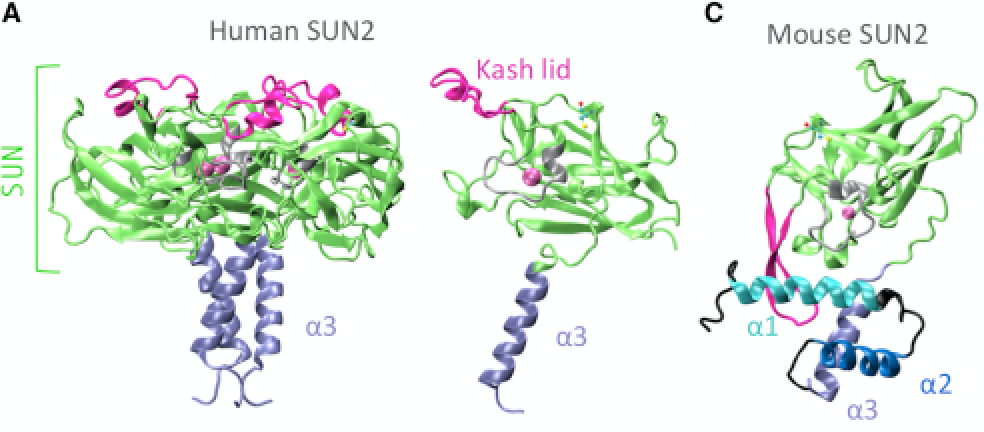
Z Jahed, D Fadavi, U Vu, E Asgari, GWG Luxton, MRK Mofrad (2018), Molecular Insights into the Mechanisms of SUN1 Oligomerization in the Nuclear Envelope. Biophysical Journal.
The LINC complex is found in a wide variety of organisms and is formed by the transluminal interaction between outer- and inner-nuclear-membrane KASH and SUN proteins, respectively. Most extensively studied are SUN1 and SUN2 proteins, which are widely expressed in mammals. Although SUN1 and SUN2 play functionally redundant roles in several cellular processes, more recent studies have revealed diverse and distinct functions for SUN1. While several recent in vitro structural studies have revealed the molecular details of various fragments of SUN2, no such structural information is available for SUN1. Herein, we conduct a systematic analysis of the molecular relationships between SUN1 and SUN2, highlighting key similarities and differences that could lead to clues into their distinct functions. We use a wide range of computational tools, including multiple sequence alignments, homology modeling, molecular docking, and molecular dynamic simulations, to predict structural differences between SUN1 and SUN2, with the goal of understanding the molecular mechanisms underlying SUN1 oligomerization in the nuclear envelope. Our simulations suggest that the structural model of SUN1 is stable in a trimeric state and that SUN1 trimers can associate through their SUN domains to form lateral complexes. We also ask whether SUN1 could adopt an inactive monomeric conformation as seen in SUN2. Our results imply that the KASH binding domain of SUN1 is also inhibited in monomeric SUN1 but through weaker interactions than in monomeric SUN2.
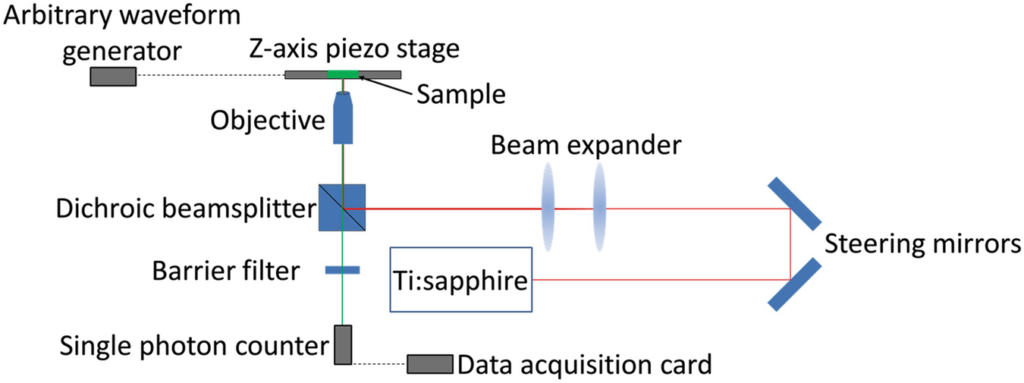
J Hennen, I Angert, KH Hur, GWG Luxton, JD Mueller (2018), Investigating LINC complex protein homo-oligomerization in the nuclear envelopes of living cells using fluorescence fluctuation spectroscopy. The LINC Complex.
Linkers of nucleoskeleton and cytoskeleton (LINC) complexes are conserved nuclear envelope (NE) spanning molecular bridges which mechanically integrate the nucleus with the cytoskeleton and mediate force transmission into the nucleoplasm. Despite their critical roles in fundamental cellular processes such as meiotic chromosome and nuclear positioning, the mechanism of LINC complex assembly in cells remains unclear. To begin to address this deficit, we recently developed z-scan fluorescence fluctuation spectroscopy (FFS) and brightness analysis as a method for quantifying the oligomeric states of fluorescent protein-tagged NE proteins including nesprins and SUN proteins. Since the homo-oligomerization of SUN2 is critical for its ability to interact with nesprins within the perinuclear space, the knowledge obtained through quantitative brightness experiments reveals important insights into the in vivo mechanisms of LINC complex assembly. Here we describe the procedure we use to determine the brightness of proteins in the NE of living cells. In addition to the measurement procedure, we discuss the instrumentation requirements and present the results of applying this procedure to measure the brightness of nesprin-2 and SUN2.
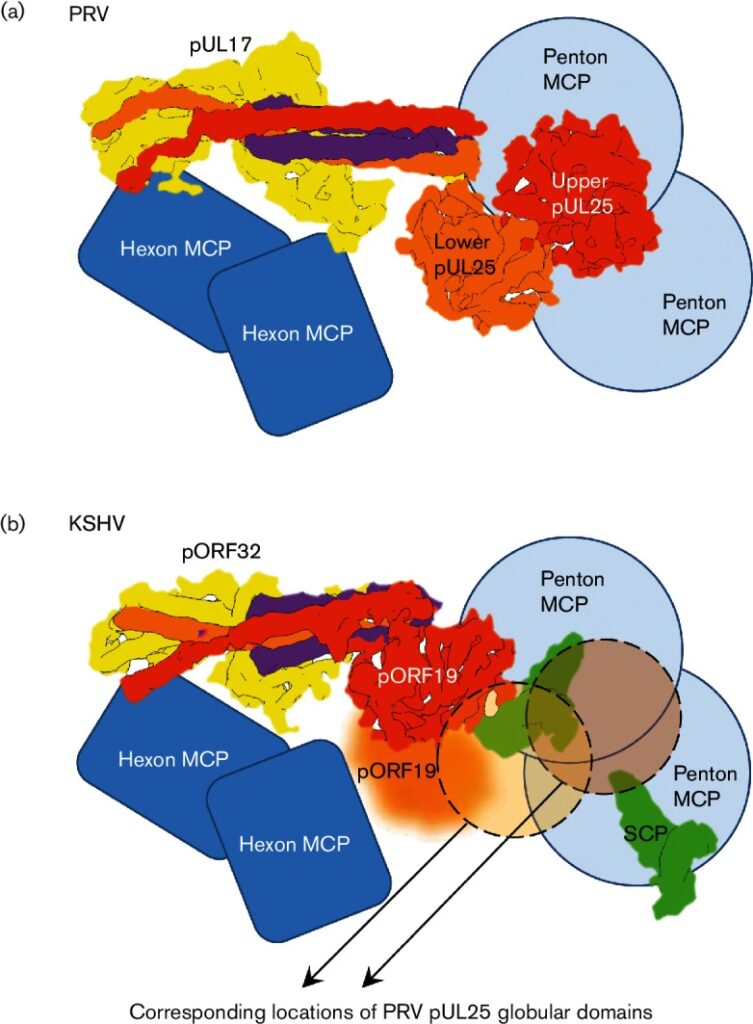
YT Liu, J Jiang, KP Bohannon, X Dai, GWG Luxton, WH Hui, GQ Bi, GA Smith, ZH Zhou (2017), A pUL25 dimer interfaces the pseudorabies virus capsid and tegument. Journal of General Virology.
Inside the virions of α-herpesviruses, tegument protein pUL25 anchors the tegument to capsid vertices through direct interactions with tegument proteins pUL17 and pUL36. In addition to promoting virion assembly, both pUL25 and pUL36 are critical for intracellular microtubule-dependent capsid transport. Despite these essential roles during infection, the stoichiometry and precise organization of pUL25 and pUL36 on the capsid surface remain controversial due to the insufficient resolution of existing reconstructions from cryo-electron microscopy (cryoEM). Here, we report a three-dimensional (3D) icosahedral reconstruction of pseudorabies virus (PRV), a varicellovirus of the α-herpesvirinae subfamily, obtained by electron-counting cryoEM at 4.9 Å resolution. Our reconstruction resolves a dimer of pUL25 forming a capsid-associated tegument complex with pUL36 and pUL17 through a coiled coil helix bundle, thus correcting previous misinterpretations. A comparison between reconstructions of PRV and the γ-herpesvirus Kaposi’s sarcoma-associated herpesvirus (KSHV) reinforces their similar architectures and establishes important subfamily differences in the capsid-tegument interface.
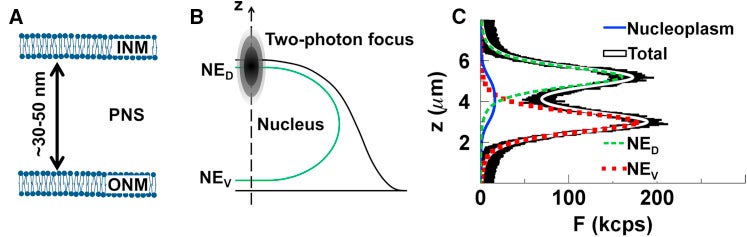
J Hennen, KH Hur, CA Saunders, GWG Luxton, JD Mueller (2017), Quantitative brightness analysis of protein oligomerization in the nuclear envelope. Biophysical Journal.
Brightness analysis of fluorescence fluctuation experiments has been used to successfully measure the oligomeric state of proteins at the plasma membrane, in the nucleoplasm, and in the cytoplasm of living cells. Here we extend brightness analysis to the nuclear envelope (NE), a double membrane barrier separating the cytoplasm from the nucleoplasm. Results obtained by applying conventional brightness analysis to fluorescently tagged proteins within the NE exhibited an unusual concentration dependence. Similarly, the autocorrelation function of the fluorescence fluctuations exhibited unexpected changes with protein concentration. These observations motivated the application of mean-segmented Q analysis, which identified the existence of a fluctuation process distinct from molecular diffusion in the NE. We propose that small changes in the separation of the inner and outer nuclear membrane are responsible for the additional fluctuation process, as suggested by results obtained for luminal and nuclear membrane-associated EGFP-tagged proteins. Finally, we applied these insights to study the oligomerization of the luminal domains of two nuclear membrane proteins, nesprin-2 and SUN2, which interact transluminally to form a nuclear envelope-spanning linker molecular bridge known as the linker of the nucleoskeleton and cytoskeleton complex.
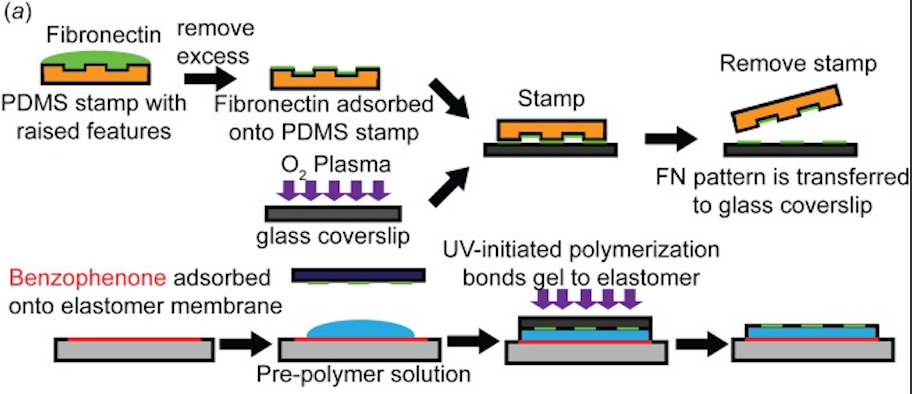
Z Win, JM Buksa, KE Steucke, GWG Luxton, VH Barocas, PW Alford (2017), Cellular Microbiaxial Stretching to Measure a Single-Cell Strain Energy Density Function. Journal of Biomechanical Engineering.
The stress in a cell due to extracellular mechanical stimulus is determined by its mechanical properties, and the structural organization of many adherent cells suggests that their properties are anisotropic. This anisotropy may significantly influence the cells’ mechanotransductive response to complex loads, and has important implications for development of accurate models of tissue biomechanics. Standard methods for measuring cellular mechanics report linear moduli that cannot capture large-deformation anisotropic properties, which in a continuum mechanics framework are best described by a strain energy density function (SED). In tissues, the SED is most robustly measured using biaxial testing. Here, we describe a cellular microbiaxial stretching (CμBS) method that modifies this tissue-scale approach to measure the anisotropic elastic behavior of individual vascular smooth muscle cells (VSMCs) with nativelike cytoarchitecture. Using CμBS, we reveal that VSMCs are highly anisotropic under large deformations. We then characterize a Holzapfel-Gasser-Ogden type SED for individual VSMCs and find that architecture-dependent properties of the cells can be robustly described using a formulation solely based on the organization of their actin cytoskeleton. These results suggest that cellular anisotropy should be considered when developing biomechanical models, and could play an important role in cellular mechano-adaptation.
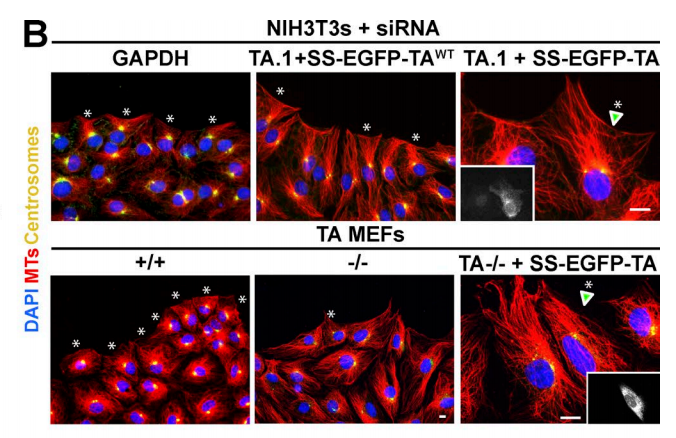
CA Saunders, NJ Harris, PT Willey, BM Woolums, Y Wang, AJ McQuown, A Schoenhofen, HJ Worman, WT Dauer, GG Gundersen*, GWG Luxton* (2017), TorsinA controls TAN line assembly and the retrograde flow of dorsal perinuclear actin cables during rearward nuclear movement. Journal of Cell Biology. (*Equal contribution)
The nucleus is positioned toward the rear of most migratory cells. In fibroblasts and myoblasts polarizing for migration, retrograde actin flow moves the nucleus rearward, resulting in the orientation of the centrosome in the direction of migration. In this study, we report that the nuclear envelope-localized AAA+ (ATPase associated with various cellular activities) torsinA (TA) and its activator, the inner nuclear membrane protein lamina-associated polypeptide 1 (LAP1), are required for rearward nuclear movement during centrosome orientation in migrating fibroblasts. Both TA and LAP1 contributed to the assembly of transmembrane actin-associated nuclear (TAN) lines, which couple the nucleus to dorsal perinuclear actin cables undergoing retrograde flow. In addition, TA localized to TAN lines and was necessary for the proper mobility of EGFP-mini-nesprin-2G, a functional TAN line reporter construct, within the nuclear envelope. Furthermore, TA and LAP1 were indispensable for the retrograde flow of dorsal perinuclear actin cables, supporting the recently proposed function for the nucleus in spatially organizing actin flow and cytoplasmic polarity. Collectively, these results identify TA as a key regulator of actin-dependent rearward nuclear movement during centrosome orientation.
KJ Petersen, HV Goodson, AL Arthur, GWG Luxton, A Houdusse, MA Titus. (2016), MyTH4-FERM myosins have an ancient and conserved role in filopod formation. Proceedings of the National Academy of Sciences.
The formation of filopodia in Metazoa and Amoebozoa requires the activity of myosin 10 (Myo10) in mammalian cells and of Dictyostelium unconventional myosin 7 (DdMyo7) in the social amoeba Dictyostelium. However, the exact roles of these MyTH4-FERM myosins (myosin tail homology 4-band 4.1, ezrin, radixin, moesin; MF) in the initiation and elongation of filopodia are not well defined and may reflect conserved functions among phylogenetically diverse MF myosins. Phylogenetic analysis of MF myosin domains suggests that a single ancestral MF myosin existed with a structure similar to DdMyo7, which has two MF domains, and that subsequent duplications in the metazoan lineage produced its functional homolog Myo10. The essential functional features of the DdMyo7 myosin were identified using quantitative live-cell imaging to characterize the ability of various mutants to rescue filopod formation in myo7-null cells. The two MF domains were found to function redundantly in filopod formation with the C-terminal FERM domain regulating both the number of filopodia and their elongation velocity. DdMyo7 mutants consisting solely of the motor plus a single MyTH4 domain were found to be capable of rescuing the formation of filopodia, establishing the minimal elements necessary for the function of this myosin. Interestingly, a chimeric myosin with the Myo10 MF domain fused to the DdMyo7 motor also was capable of rescuing filopod formation in the myo7-null mutant, supporting fundamental functional conservation between these two distant myosins. Together, these findings reveal that MF myosins have an ancient and conserved role in filopod formation.
C Coombes, A Yamamoto, M McClellan, TA Reid, M Plooster, GWG Luxton, J Alper, J Howard, MK Gardner. (2016), Mechanism of microtubule lumen entry for the α-tubulin acetyltransferase enzyme αTAT1. Proceedings of the National Academy of Sciences.
Microtubules are structural polymers inside of cells that are subject to posttranslational modifications. These posttranslational modifications create functionally distinct subsets of microtubule networks in the cell, and acetylation is the only modification that takes place in the hollow lumen of the microtubule. Although it is known that the α-tubulin acetyltransferase (αTAT1) is the primary enzyme responsible for microtubule acetylation, the mechanism for how αTAT1 enters the microtubule lumen to access its acetylation sites is not well understood. By performing biochemical assays, fluorescence and electron microscopy experiments, and computational simulations, we found that αTAT1 enters the microtubule lumen through the microtubule ends, and through bends or breaks in the lattice. Thus, microtubule structure is an important determinant in the acetylation process. In addition, once αTAT1 enters the microtubule lumen, the mobility of αTAT1 within the lumen is controlled by the affinity of αTAT1 for its acetylation sites, due to the rapid rebinding of αTAT1 onto highly concentrated α-tubulin acetylation sites. These results have important implications for how acetylation could gradually accumulate on stable subsets of microtubules inside of the cell.
KN Dahl, GWG Luxton (2016), A Special Topic on Nuclear Mechanobiology. Cellular and Molecular Bioengineering.
The view of the cell nucleus has evolved from an isolated, static organelle to a dynamic structure integrated with other mechanical elements of the cell. Both dynamics and integration appear to contribute to a mechanical regulation of genome expression. Here, we review physical structures inside the nucleus at different length scales and the dynamic reorganization modulated by cellular forces. First, we discuss nuclear organization focusing on self-assembly and disassembly of DNA structures and various nuclear bodies. We then discuss the importance of connections from the chromatin fiber through the nuclear envelope to the rest of the cell as they relate to mechanobiology. Finally, we discuss how cell stimulation, both chemical and physical, can alter nuclear structures and ultimately cellular function in healthy cells and in some model diseases. The view of chromatin and nuclear bodies mechanical entities integrated with force generation from the cytoskeleton combines polymer physics with cell biology and medicine.
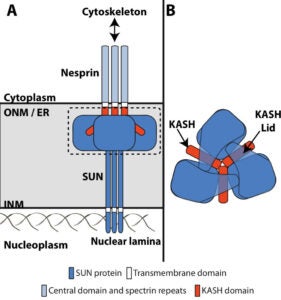
CA Saunders, GWG Luxton (2016), LINCing defective nuclear-cytoskeletal coupling and DYT1 dystonia. Cellular and Molecular Bioengineering.
Mechanical forces generated by nuclear-cytoskeletal coupling through the LINC (linker of nucleoskeleton and cytoskeleton) complex, an evolutionarily conserved molecular bridge in the nuclear envelope (NE), are critical for the execution of wholesale nuclear positioning events in migrating and dividing cells, chromosome dynamics during meiosis, and mechanotransduction. LINC complexes consist of outer (KASH (Klarsicht, ANC-1, and Syne homology)) and inner (SUN (Sad1, UNC-84)) nuclear membrane proteins. KASH proteins interact with the cytoskeleton in the cytoplasm and SUN proteins in the perinuclear space of the NE. In the nucleoplasm, SUN proteins interact with A-type nuclear lamins and chromatin-binding proteins. Recent structural insights into the KASH-SUN interaction have generated several questions regarding how LINC complex assembly and function might be regulated within the perinuclear space. Here we discuss potential LINC regulatory mechanisms and focus on the potential role of AAA+ (ATPases associated with various cellular activities) protein, torsinA, as a LINC complex regulator within the NE. We also examine how defects in LINC complex regulation by torsinA may contribute to the pathogenesis of the human neurological movement disorder, DYT1 dystonia.
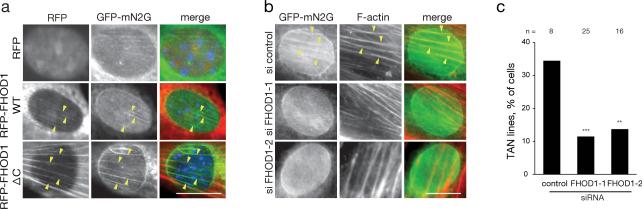
S Kutscheidt, R Zhu, S Antoku, GWG Luxton, I Stagljar, OT Fackler, GG Gundersen. (2014), FHOD1 interaction with nesprin-2G mediates TAN line formation and nuclear movement. Nature Cell Biology.
Active positioning of the nucleus is integral to division, migration, and differentiation of mammalian cells1. Fibroblasts polarizing for migration orient their centrosomes by actin-dependent nuclear movement2. This nuclear movement depends on nesprin-2 giant (N2G), a large, actin-binding outer nuclear membrane component of transmembrane actin-associated (TAN) lines that couple nuclei to moving actin cables3. Here, we identify the diaphanous formin FHOD1 as an interaction partner of N2G. Silencing FHOD1 expression or expression of fragments containing binding sites of N2G or FHOD1 disrupted nuclear movement and centrosome orientation in polarizing fibroblasts. Unexpectedly, silencing of FHOD1 expression did not affect the formation or rearward flow of dorsal actin cables required for nuclear positioning. Rather, N2G-FHOD1 interaction provided a second connection to actin cables essential for TAN line formation and thus nuclear movement. These results reveal a unique function for a formin in coupling an organelle to actin filaments for translocation and suggest that TAN lines require multi-point attachments to actin cables to resist the large forces necessary to move the nucleus.

GWG Luxton*, DA Starr* (2014), KASHing up with the nucleus: novel functional roles of KASH proteins at the cytoplasmic surface of the nucleus. Current Opinion in Cell Biology. (*Equal contribution)
Nuclear-cytoskeletal connections are central to fundamental cellular processes, including nuclear positioning and chromosome movements in meiosis. The cytoskeleton is coupled to the nucleoskeleton through conserved KASH-SUN bridges, or LINC complexes, that span the nuclear envelope. KASH proteins localize to the outer nuclear membrane where they connect the nucleus to the cytoskeleton. New findings have expanded the functional diversity of KASH proteins, showing that they interact with microtubule motors, actin, intermediate filaments, a nonconventional myosin, RanGAP, and each other. The role of KASH proteins in cellular mechanics is discussed. Genetic mutations in KASH proteins are associated with autism, hearing loss, cancer, muscular dystrophy and other diseases.
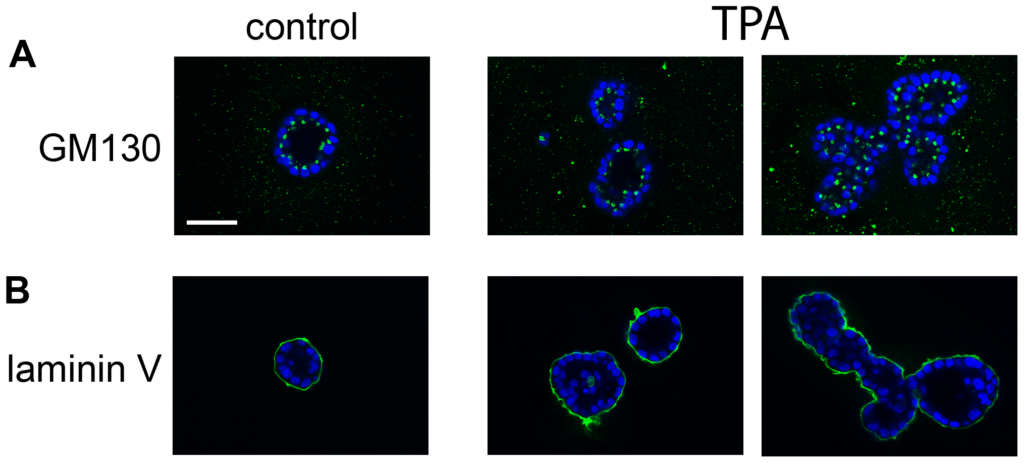
KS Klos, JK Warmka, DM Drachenberg, L Chang, GWG Luxton, CT Leung, KL Schwertfeger, EV Wattenberg (2014), Building bridges toward invasion: tumor promoter treatment induces a novel protein kinase C-dependent phenotype in MCF10A mammary cell acini. Plos One.
The potent tumor promoter 12-O-tetradecanoylphorbol-13-acetate (TPA) alters many cellular processes through activation of its receptor protein kinase C (PKC), including gene expression, cell cycle, and the regulation of cell morphology, raising an important question for developing targeted methods to prevent cancer: which effects of TPA are crucial for carcinogenesis? To address this question, we studied TPA action in the 3-dimensional (3D) MCF10A human breast epithelial cell system, which models important features of in vivo epithelial tissue including growth constraints, structural organization of cells, and establishment of a basement membrane. MCF10A cells, which are immortalized but nontumorigenic, form hollow, spheroid structures in 3D culture referred to as acini. The development of normal acini requires the tight spatiotemporal regulation of cellular proliferation, polarization, apoptosis, and growth arrest. Treatment of MCF10A acini with TPA caused the appearance of multi-acinar structures. Surprisingly, this phenotype did not involve an increase in cell number or major changes in cell death, and polarization. Instead, live cell and confocal microscopy revealed that TPA stimulates MCF10A acini to aggregate. TPA induces the PKC-dependent production of actin-based protrusions, which leads to the formation of cellular bridges between acini, the clustering of acini, and allows cells to move into adjacent acini. During this process, the integrity of the laminin V basement membrane is disrupted, while E-cadherin-based cell-cell contacts remain intact. Altogether, our results show that under the biochemical and structural constraints of epithelial tissue, as modeled by the 3D MCF10A system, TPA induces a novel PKC-dependent phenotype that resembles local invasion. Of the many effects caused by TPA, these studies highlight the aggressive production of actin-based cellular protrusions as a potentially important event along the pathway to carcinogenesis.
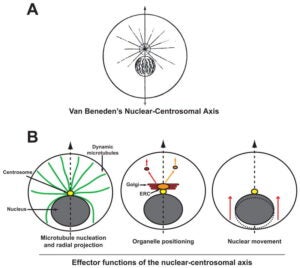
GWG Luxton, GG Gundersen (2011), Orientation and function of the nuclear–centrosomal axis during cell migration. Current Opinion in Cell Biology.
A hallmark of polarity in most migrating cells is the orientation of the nuclear centrosomal (NC) axis relative to the front-back cellular axis. Here, we review ‘effector functions’ associated with the NC axis during cell migration. We highlight recent research that has demonstrated that the orientation of the NC axis depends upon the coordinated, but separate positioning of the nucleus and the centrosome. We stress the importance of environmental factors such as cell-cell contacts and substrate topology for NC axis orientation. Finally, we summarize tests of the significance of this axis for cell migration and disease.
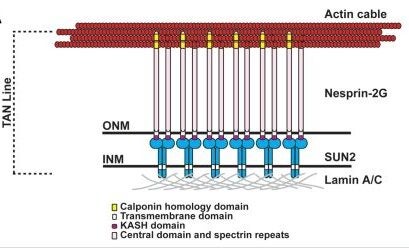
GWG Luxton, ER Gomes, ES Folker, H Worman, GG Gundersen (2011), TAN lines: a novel nuclear envelope structure involved in nuclear positioning. Nucleus.
Nuclear position is actively controlled and can be adjusted according to the needs of a cell by nuclear movement. Microtubules mediate the majority of nuclear movements studied to date, although examples of nuclear movements mediated by the actin cytoskeleton have been described. One such actin-dependent nuclear movement occurs during centrosome orientation in fibroblasts polarizing for migration. Here, the centrosome is maintained at the cell center while the nucleus is moved to the cell rear by actin retrograde flow thus positioning the centrosome between the nucleus and the leading edge of the cell. We have explored the molecular mechanism for actin dependent movement of the nucleus during centrosome centration. We found that a novel linear array of nuclear envelope membrane proteins composed of nesprin-2G and SUN2, called transmembrane actin-associated nuclear (TAN) lines, couple the nucleus to moving actin cables resulting in the nucleus being positioned toward the cell rear. TAN lines are anchored by A-type lamins and this allows the forces generated by the actin cytoskeleton to be transmitted across the nuclear envelope to move the nucleus. Here we review the data supporting this mechanism for nuclear movement, discuss questions remaining to be addressed and consider how this new mechanism of nuclear movement may shed light on human disease.
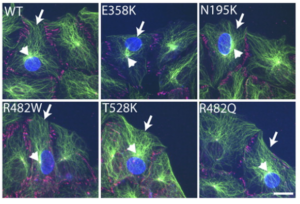
ES Folker, C Östlund, GWG Luxton, HJ Worman, GG Gundersen (2010), Lamin A variants that cause striated muscle disease are defective in anchoring transmembrane actin-associated nuclear lines for nuclear movement. Proceedings of the National Academy of Sciences.
Mutations in LMNA, which encodes A-type lamins, result in disparate diseases, known collectively as laminopathies, that affect distinct tissues, including striated muscle and adipose tissue. Lamins provide structural support for the nucleus and sites of attachment for chromatin, and defects in these functions may contribute to disease pathogenesis. Recent studies suggest that A-type lamins may facilitate connections between the nucleus and the cytoskeleton mediated by nuclear envelope nesprin and SUN proteins. In mammalian cells, however, interfering with A-type lamins does not affect the localization of these proteins. Here, we used centrosome orientation in fibroblasts, which requires separate nuclear and centrosome positioning pathways, as a model system to understand how LMNA mutations affect nucleus-cytoskeletal connections. We find that LMNA mutations causing striated muscle diseases block actin-dependent nuclear movement, whereas most that affect adipose tissue inhibit microtubule-dependent centrosome positioning. Genetic deletion or transient depletion of A-type lamins also blocked nuclear movement, showing that mutations affecting muscle exhibit the null phenotype. Lack of A-type lamins, or expression of variants that cause striated muscle disease, did not affect assembly of nesprin-2G and SUN2 into transmembrane actin-associated nuclear (TAN) lines that attach the nucleus to retrogradely moving actin cables. Nesprin-2G TAN lines were less stable, however, and slipped over the nucleus rather than moving with it, indicating that they were not anchored. Nesprin-2G TAN lines also slipped in SUN2-depleted cells. Our results establish A-type lamins as anchors for nesprin-2G–SUN2 TAN lines to allow productive movement and proper positioning of the nucleus by actin.
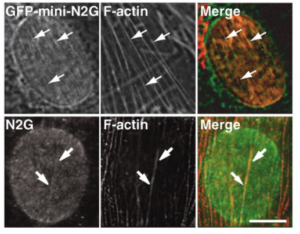
GWG Luxton*, ER Gomes*, ES Folker, E Vintinner, GG Gundersen (2010), Linear arrays of nuclear envelope proteins harness retrograde actin flow for nuclear movement. Science. (*Equal contribution)
Nuclei move to specific locations to polarize migrating and differentiating cells. Many nuclear movements are microtubule-dependent. However, nuclear movement to reorient the centrosome in migrating fibroblasts occurs through an unknown actin-dependent mechanism. We found that linear arrays of outer (nesprin2G) and inner (SUN2) nuclear membrane proteins assembled on and moved with retrogradely moving dorsal actin cables during nuclear movement in polarizing fibroblasts. Inhibition of nesprin2G, SUN2, or actin prevented nuclear movement and centrosome reorientation. The coupling of actin cables to the nuclear membrane for nuclear movement via specific membrane proteins indicates that, like plasma membrane integrins, nuclear membrane proteins assemble into actin-dependent arrays for force transduction.
GWG Luxton, GG Gundersen (2007), HDAC6-pack: cortactin acetylation joins the brew. Developmental Cell.
The reversible acetylation of lysine residues is an important posttranslational modification for the regulation of histones, transcription factors, chaperones, and microtubules. In a recent article in Molecular Cell, Zhang et al. (2007) describe a new target of reversible acetylation, the actin binding protein cortactin.
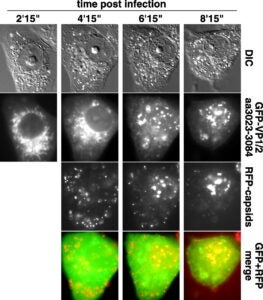
JI Lee*, GWG Luxton*, GA Smith (2006), Identification of an essential domain in the herpesvirus VP1/2 tegument protein: the carboxy terminus directs incorporation into capsid assemblons. Journal of Virology. (*Equal contribution)
The herpesvirus tegument is a layer of viral and cellular proteins located between the capsid and envelope of the virion. The VP1/2 tegument protein is critical for the propagation of all herpesviruses examined. Using an infectious clone of the alphaherpesvirus pseudorabies virus, we have made a collection of truncation and in-frame deletion mutations within the VP1/2 gene (UL36) and examined the resulting viruses for spread between cells. We found that the majority of the VP1/2 protein either was essential for virus propagation or did not tolerate large deletions. A recently described amino-terminal deubiquitinase-encoding domain was dispensable for alphaherpesvirus propagation, but the rate of propagation in an epithelial cell line and the frequency of transport in axons of primary sensory neurons were both reduced. We mapped one essential domain to a conserved sequence at the VP1/2 carboxy terminus and demonstrated that this domain sufficient to redirect the green fluorescent protein to capsid assemblons in nuclei of infected cells.
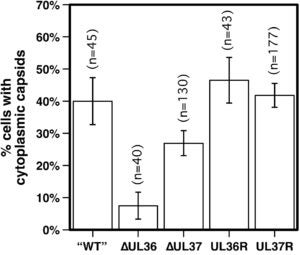
GWG Luxton*, JI Lee*, S Haverlock-Moyns, JM Schober, GA Smith (2006), The pseudorabies virus VP1/2 tegument protein is required for intracellular capsid transport. Journal of Virology. (*Equal contribution)
Transport of capsids in cells is critical to alphaherpesvirus infection and pathogenesis; however, viral factors required for transport have yet to be identified. Here we provide a detailed examination of capsid dynamics during the egress phase of infection in Vero cells infected with pseudorabies virus. We demonstrate that the VP1/2 tegument protein is required for processive microtubule-based transport of capsids in the cytoplasm. A second tegument protein that binds to VP1/2, UL37, was necessary for wild-type transport but was not essential for this process. Both proteins were also required for efficient nuclear egress of capsids to the cytoplasm.
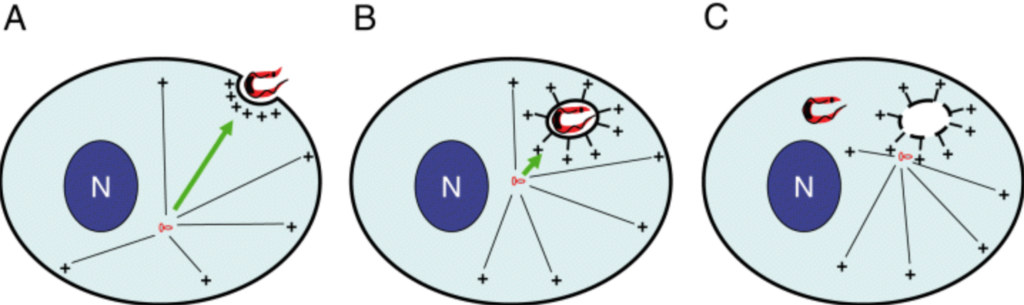
KM Tyler, GWG Luxton, DA Applewhite, SC Murphy, DM Engman (2005), Responsive microtubule dynamics promote cell invasion by Trypanosoma cruzi. Cellular Microbiology.
The American trypanosome, Trypanosoma cruzi, can invade non‐phagocytic cell types by a G‐protein‐mediated, calcium‐dependent mechanism, in which the cell’s natural puncture repair mechanism is usurped in order to recruit lysosomes to the parasite/host cell junction or ‘parasite synapse.’ The fusion of lysosomes necessary for construction of the nascent parasitophorous vacuole is achieved by directed trafficking along microtubules. We demonstrate altered host cell microtubule dynamics during the initial stages of the entry process involving de novo microtubule polymerization from the cytoplasmic face of the parasite synapse which appears to serve as a secondary microtubule organizing centre. The net result of these dynamic changes to the host cell’s microtubule cytoskeleton is the development of the necessary infrastructure for transport of lysosomes to the parasite synapse.
GWG Luxton, S Haverlock, KE Coller, SE Antinone, A Pincetic, GA Smith (2005), Targeting of herpesvirus capsid transport in axons is coupled to association with specific sets of tegument proteins. Proceedings of the National Academy of Sciences.
The capsids of neurotropic herpesviruses have the remarkable ability to move in specific directions within axons. By modulating bidirectional capsid transport to favor either retrograde (minus-end) or anterograde (plus-end) motion, these viruses travel to sensory ganglia or peripheral tissue at specific stages of infection. By using correlative motion analysis to simultaneously monitor the trafficking of distinct viral proteins in living neurons, we demonstrate that viral “tegument” proteins are complexed to capsids moving in axons. The removal of a subset of tegument proteins from capsids invariably preceded retrograde transport to the cell body in sensory ganglia, whereas addition of these proteins was coupled to anterograde transport of progeny capsids to the distal axon. Although capsid transport never occurred without associated tegument proteins, anterograde-specific tegument proteins were competent to travel to the distal axon independent of capsids. These findings are compatible with a model of viral bidirectional transport in which tegument proteins direct capsid traffic to specific intracellular locations during the infectious cycle.
Z Yan, R Zak, GWG Luxton, TC Ritchie, U Bantel-Schaal, JF Engelhardt (2002), Ubiquitination of both adeno-associated virus type 2 and 5 capsid proteins affects the transduction efficiency of recombinant vectors. Journal of Virology.
In the presence of complementing adeno-associated virus type 2 (AAV-2) Rep proteins, AAV-2 genomes can be pseudotyped with the AAV-5 capsid to assemble infectious virions. Using this pseudotyping strategy, the involvement of the ubiquitin-proteasome system in AAV-5 and AAV-2 capsid-mediated infections was compared. A recombinant AAV-2 (rAAV-2) proviral luciferase construct was packaged into both AAV-2 and AAV-5 capsid particles, and transduction efficiencies in a number of cell lines were compared. Using luciferase expression as the end point, we demonstrated that coadministration of the viruses with proteasome inhibitors not only increased the transduction efficiency of rAAV-2, as previously reported, but also augmented rAAV-5-mediated gene transfer. Increased transgene expression was independent of viral genome stability, since there was no significant difference in the amounts of internalized viral DNA in the presence or absence of proteasome inhibitors. Western blot assays of immunoprecipitated viral capsid proteins from infected HeLa cell lysates and in vitro reconstitution experiments revealed evidence for ubiquitin conjugation of both AAV-2 and AAV-5 capsids. Interestingly, heat-denatured virus particles were preferential substrates for in vitro ubiquitination, suggesting that endosomal processing of the viral capsid proteins is a prelude to ubiquitination. Furthermore, ubiquitination may be a signal for processing of the capsid at the time of virion disassembly. These studies suggest that the previously reported influences of the ubiquitin-proteasome system on rAAV-2 transduction are also active for rAAV-5 and provide a clearer mechanistic framework for understanding the functional significance of ubiquitination.