Full List of Publications: Google Scholar
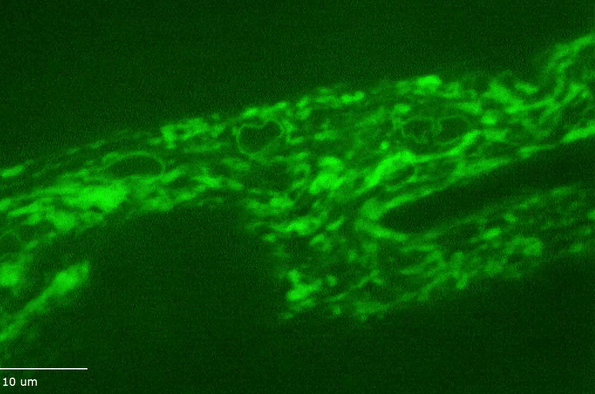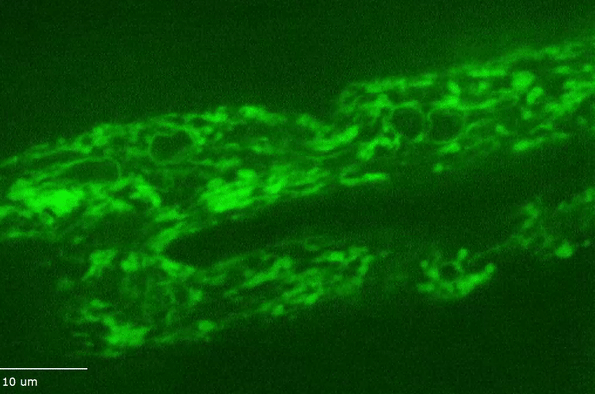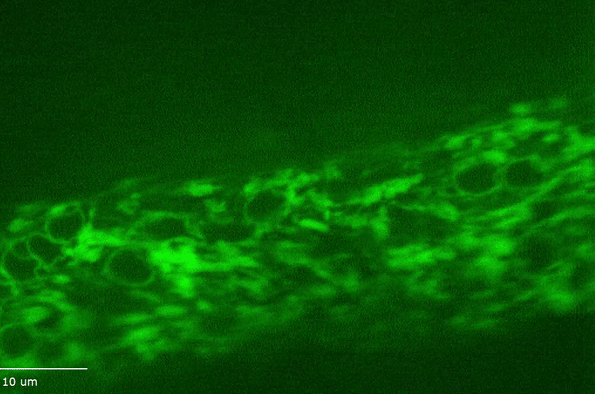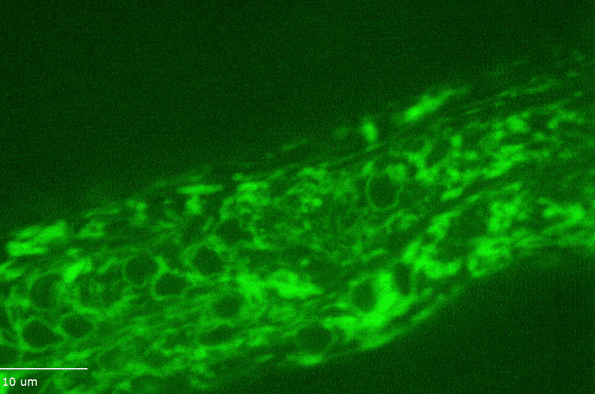
H Hao, S Kalra, LE Jameson, LA Guerrero, NE Cain, J Bolivar, DA Starr (2021), The Nesprin-1/-2 ortholog ANC-1 regulates organelle positioning in C. elegans independently from its KASH or actin-binding domains. eLife.
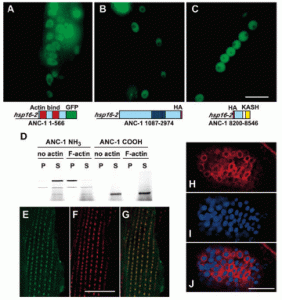
KASH proteins in the outer nuclear membrane comprise the cytoplasmic half of LINC complexes that connect nuclei to the cytoskeleton. Caenorhabditis elegans ANC-1, an ortholog of Nesprin-1/2, contains actin-binding and KASH domains at opposite ends of a long spectrin-like region. Deletion of either the KASH or calponin homology (CH) domains does not completely disrupt nuclear positioning, suggesting neither KASH nor CH domains are essential. Deletions in the spectrin-like region of ANC-1 led to significant defects, but only recapitulated the null phenotype in combination with mutations in the trans-membrane span. In anc-1 mutants, the ER was unanchored, moving throughout the cytoplasm, and often fragmented. The data presented here support a cytoplasmic integrity model where ANC-1 localizes to the ER membrane and extends into the cytoplasm to position nuclei, ER, mitochondria, and likely other organelles in place.
L Ma, DA Starr (2020). Membrane fusion drives pronuclear meeting in the one-cell embryo. Invited Perspective in The Journal of Cell Biology.
The mechanisms that control how the two parental pronuclei fuse in the first mitosis of the embryo are poorly understood. In this issue, Rahman et al. (2020. J. Cell Biol.) found that membrane fusion between pronuclear envelopes, followed by fenestration, promotes pronuclear fusion.
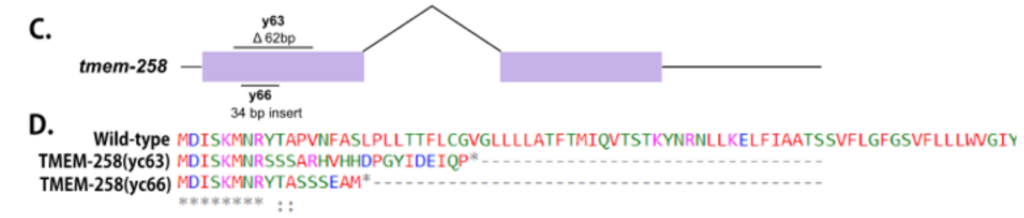
EF Gregory, DA Starr (2020). tmem-258 is dispensable for both nuclear anchorage and migration in C. elegans. Micropublication:biology.
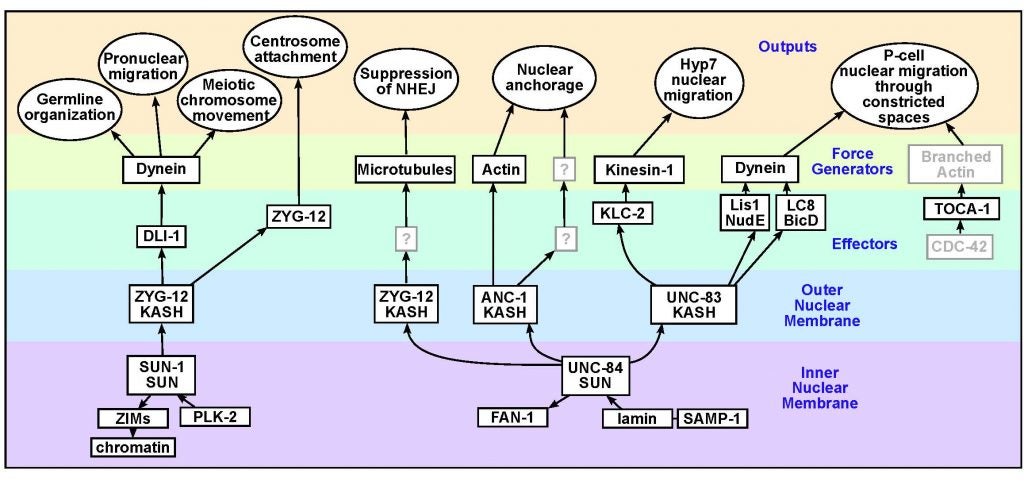
DA Starr (2019), A network of nuclear envelope proteins and cytoskeletal force generators mediates movements of and within nuclei throughout C. elegans development. Experimental Biology and Medicine.
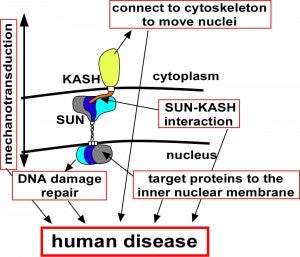
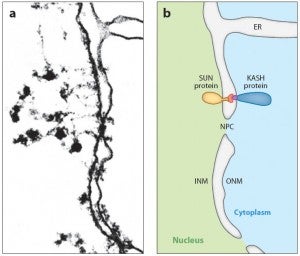
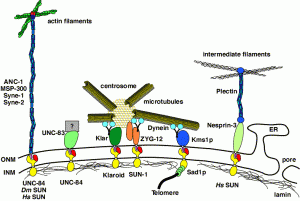
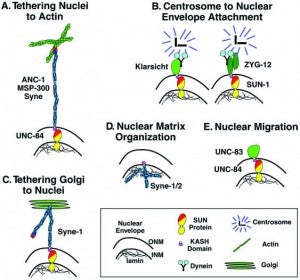
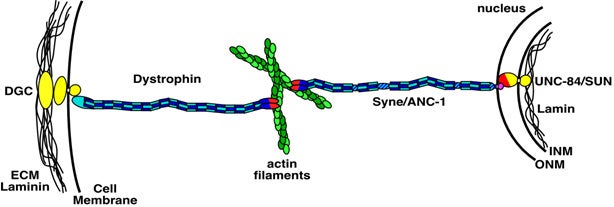
Nuclear migration and anchorage, together referred to as nuclear positioning, are central to many cellular and developmental events. Nuclear positioning is mediated by a conserved network of nuclear envelope proteins that interacts with force generators in the cytoskeleton. At the heart of this network are LINC (linker of nucleoskeleton and cytoskeleton) complexes made of SUN (Sad1 and UNC-84) proteins at the inner nuclear membrane and KASH (Klarsicht, ANC-1, and Syne homology) proteins in the outer nuclear membrane. LINC complexes span the nuclear envelope, maintain nuclear envelope architecture, designate the surface of nuclei distinctly from the contiguous ER, and were instrumental in the early evolution of eukaryotes. LINC complexes interact with lamins in the nucleus and with various cytoplasmic KASH effectors from the surface of nuclei. These effectors regulate the cytoskeleton, leading to a variety of cellular outputs including pronuclear migration, nuclear migration through constricted spaces, nuclear anchorage, centrosome attachment to nuclei, meiotic chromosome movements, and DNA damage repair. How LINC complexes are regulated and how they function are reviewed here. The focus is on recent studies elucidating the best-understood network of LINC complexes, those used throughout C. elegans development.
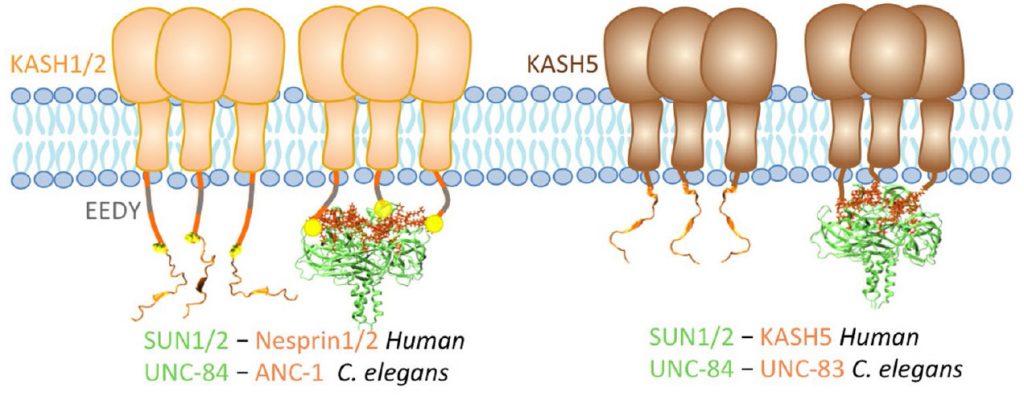
Role of KASH lengths in the regulation of LINC complexes. Molecular Biology of the Cell.
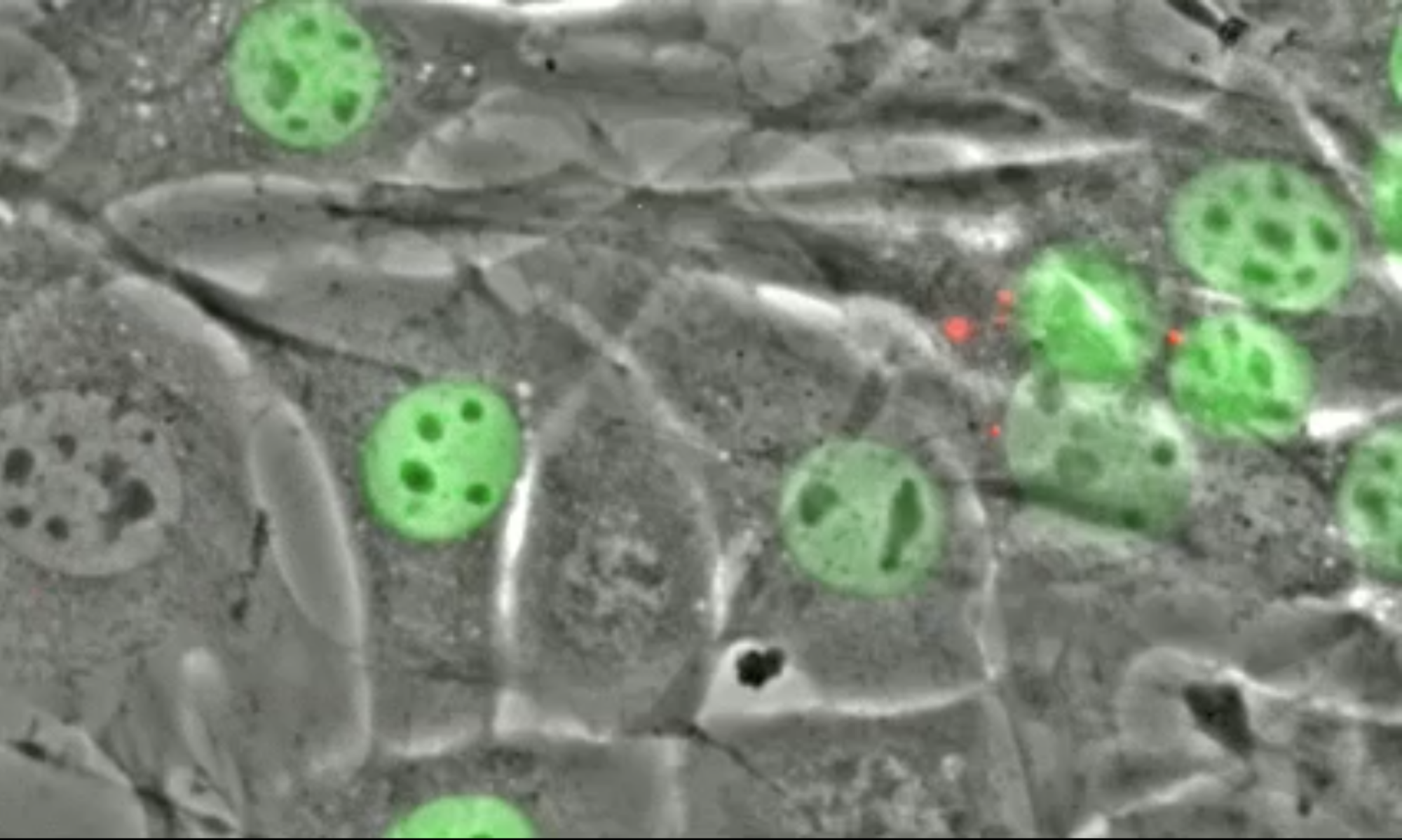
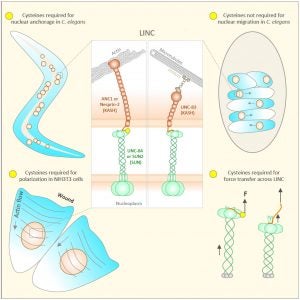
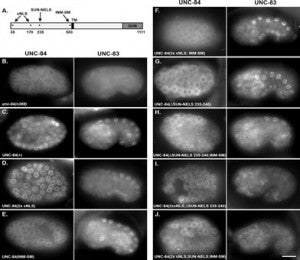
The linker of the nucleoskeleton and cytoskeleton (LINC) complex is formed by the conserved interactions between Sad-1 and UNC-84 (SUN) and Klarsicht, ANC-1, SYNE homology (KASH) domain proteins, providing a physical coupling between the nucleoskeleton and cytoskeleton that mediates the transfer of physical forces across the nuclear envelope. The LINC complex can perform distinct cellular functions by pairing various KASH domain proteins with the same SUN domain protein. For example, in Caenorhabditis elegans, SUN protein UNC-84 binds to two KASH proteins UNC-83 and ANC-1 to mediate nuclear migration and anchorage, respectively. In addition to distinct cytoplasmic domains, the luminal KASH domain also varies among KASH domain proteins of distinct functions. In this study, we combined in vivo C. elegans genetics and in silico molecular dynamics simulations to understand the relation between the length and amino acid composition of the luminal KASH domain, and the function of the SUN–KASH complex. We show that longer KASH domains can withstand and transfer higher forces and interact with the membrane through a conserved membrane proximal EEDY domain that is unique to longer KASH domains. In agreement with our models, our in vivo results show that swapping the KASH domains of ANC-1 and UNC-83, or shortening the KASH domain of ANC-1, both result in a nuclear anchorage defect in C. elegans.
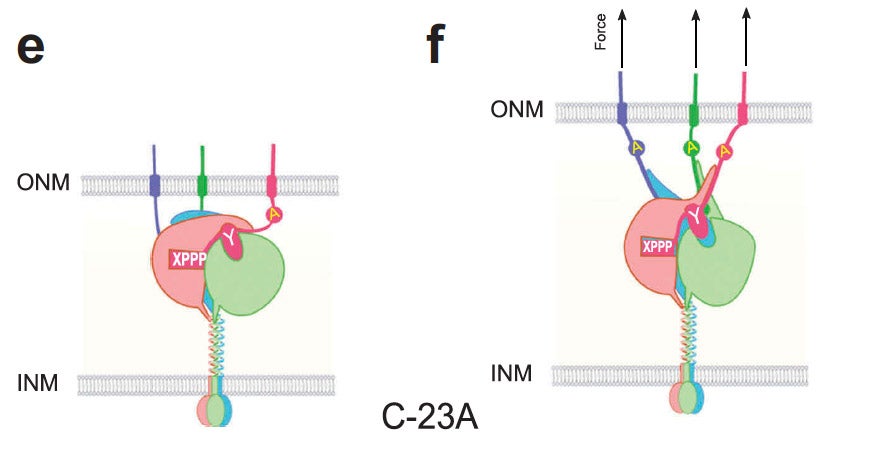
Nucleus.
LINC complexes (Linker of Nucleoskeleton and Cytoskeleton), consisting of inner nuclear membrane SUN (Sad1, UNC-84) proteins and outer nuclear membrane KASH (Klarsicht, ANC-1, and Syne Homology) proteins, are essential for nuclear positioning, cell migration and chromosome dynamics. To test the in vivo functions of conserved interfaces revealed by crystal structures, Cain et al used a combination of Caenorhabditis elegans genetics, imaging in cultured NIH 3T3 fibroblasts, and Molecular Dynamic simulations, to study SUN-KASH interactions. Conserved aromatic residues at the -7 position of the C-termini of KASH proteins and conserved disulfide bonds in LINC complexes play important roles in force transmission across the nuclear envelope. Other properties of LINC complexes, such as the helices preceding the SUN domain, the longer coiled-coils spanning the perinuclear space and higher-order organization may also function to transmit mechanical forces generated by the cytoskeleton across the nuclear envelope.
Current Biology.
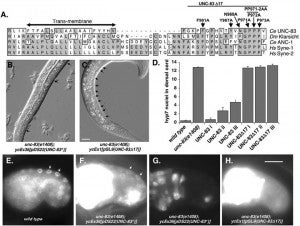
Many nuclear positioning events involve linker of nucleoskeleton and cytoskeleton (LINC) complexes, which transmit forces generated by the cytoskeleton across the nuclear envelope. LINC complexes are formed by trans-luminal interactions between inner nuclear membrane SUN proteins and outer nuclear membrane KASH proteins, but how these interactions are regulated is poorly understood. We combine in vivo C. elegans genetics, in vitro wounded fibroblast polarization, and in silico molecular dynamic simulations to elucidate mechanisms of LINC complexes. The extension of the KASH domain by a single alanine residue or the mutation of the conserved tyrosine at -7 completely blocked the nuclear migration function of C. elegans UNC-83. Analogous mutations at -7 of mouse nesprin-2 disrupted rearward nuclear movements in NIH3T3 cells, but did not disrupt ANC-1 in nuclear anchorage. Furthermore, conserved cysteines predicted to form a disulfide bond between SUN and KASH proteins are important for the function of certain LINC complexes and might promote a developmental switch between nuclear migration and nuclear anchorage. Mutations of conserved cysteines in SUN or KASH disrupted ANC-1 dependent nuclear anchorage in C. elegans and Nesprin-2G dependent nuclear movements in polarizing fibroblasts. However, the SUN cysteine mutation did not disrupt nuclear migration. Moreover, molecular dynamic simulations showed that a disulfide bond is necessary for the maximal transmission of cytoskeleton-generated forces by LINC complexes in silico. Thus, we have demonstrated functions for SUN-KASH binding interfaces, including a predicted intermolecular disulfide bond, as mechanistic determinants of nuclear positioning and may represent targets for regulation.
Unlike the classical nuclear envelope with two membranes found in other eukaryotic cells, most nematode sperm nuclei are not encapsulated by membranes. Instead, they are surrounded by a nuclear halo of unknown composition. How the halo is formed and regulated is unknown. We used forward genetics to identify molecular lesions behind three classical fer (fertilization defective) mutations that disrupt the ultrastructure of the Caenorhabditis elegans sperm nuclear halo. We found fer-2 and fer-4 alleles to be nonsense mutations in mib-1. fer-3 was caused by a nonsense mutation in eri-3. GFP::MIB-1 was expressed in the germline during early spermatogenesis, but not in mature sperm. mib-1 encodes a conserved E3 ubiquitin ligase homologous to vertebrate Mib1 and Mib2, which function in Notch signaling. Here, we show that mib-1 is important for male sterility and is involved in the regulation or formation of the nuclear halo during nematode spermatogenesis.
Unlike the classical nuclear envelope with two membranes found in other eukaryotic cells, most nematode sperm nuclei are not encapsulated by membranes. Instead, they are surrounded by a nuclear halo of unknown composition. How the halo is formed and regulated is unknown. We used forward genetics to identify molecular lesions behind three classical fer (fertilization defective) mutations that disrupt the ultrastructure of the Caenorhabditis elegans sperm nuclear halo. We found fer-2 and fer-4 alleles to be nonsense mutations in mib-1. fer-3 was caused by a nonsense mutation in eri-3. GFP::MIB-1 was expressed in the germline during early spermatogenesis, but not in mature sperm. mib-1 encodes a conserved E3 ubiquitin ligase homologous to vertebrate Mib1 and Mib2, which function in Notch signaling. Here, we show that mib-1 is important for male sterility and is involved in the regulation or formation of the nuclear halo during nematode spermatogenesis.
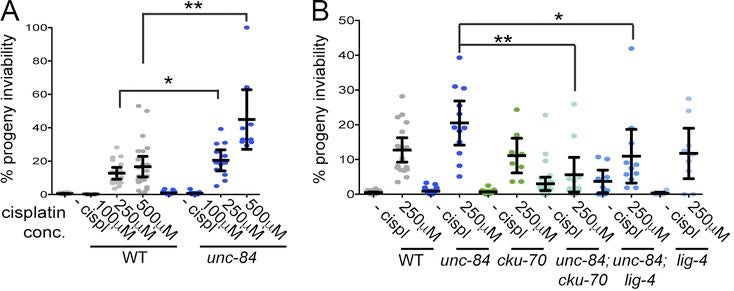
The Caenorhabditis elegans SUN domain protein, UNC-84, functions in nuclear migration and anchorage in the soma. We discovered a novel role for UNC-84 in DNA damage repair and meiotic recombination. Loss of UNC-84 leads to defects in the loading and disassembly of the recombinase RAD-51. Similar to mutations in Fanconi anemia (FA) genes, unc-84 mutants and human cells depleted of Sun-1 are sensitive to DNA cross-linking agents, and sensitivity is rescued by the inactivation of nonhomologous end joining (NHEJ). UNC-84 also recruits FA nuclease FAN-1 to the nucleoplasm, suggesting that UNC-84 both alters the extent of repair by NHEJ and promotes the processing of cross-links by FAN-1. UNC-84 interacts with the KASH protein ZYG-12 for DNA damage repair. Furthermore, the microtubule network and interaction with the nucleoskeleton are important for repair, suggesting that a functional linker of nucleoskeleton and cytoskeleton (LINC) complex is required. We propose that LINC complexes serve a conserved role in DNA repair through both the inhibition of NHEJ and the promotion of homologous recombination at sites of chromosomal breaks.
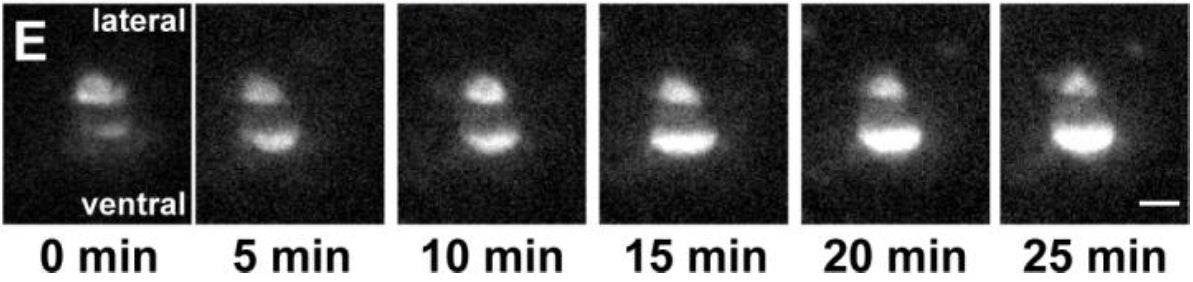
CR Bone, Y Chang, NE Cain, SP Murphy, DA Starr (2016), Nuclei migrate through constricted spaces using microtubule motors and actin networks in C. elegans hypodermal cells. Development.
Cellular migrations through constricted spaces are a critical aspect of many developmental and disease processes including hematopoiesis, inflammation, and metastasis. A limiting factor in these events is nuclear deformation. Here, we establish an in vivo model where nuclei can be visualized while moving through constrictions and use it to elucidate mechanisms for nuclear migration. C. elegans hypodermal P-cell larval nuclei traverse a narrow space about 5% their width. This constriction is blocked by fibrous organelles, structures connecting the muscles to cuticle through P cells. Fibrous organelles are removed just prior to nuclear migration, when nuclei and lamins undergo extreme morphological changes to squeeze through the space. Both actin and microtubule networks are organized to mediate nuclear migration. The LINC complex, consisting of the SUN protein UNC-84 and the KASH protein UNC-83, recruits dynein and kinesin-1 to the nuclear surface. Both motors function in P-cell nuclear migration, but dynein, functioning through UNC-83, plays a more central role as nuclei migrate toward minus ends of polarized microtubule networks. Thus, the nucleoskeleton and cytoskeleton are coordinated to move nuclei through constricted spaces.
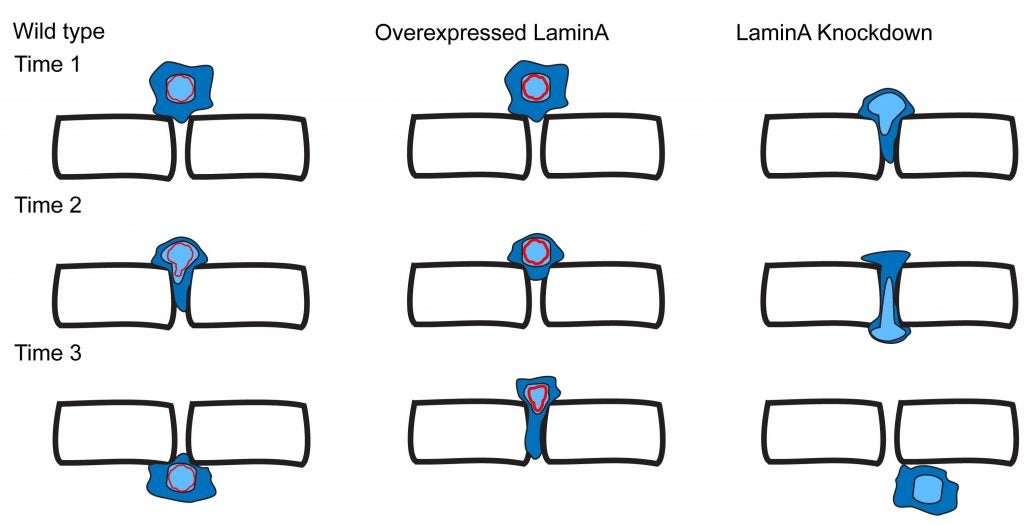
CR Bone, DA Starr (2016), Nuclear migration events throughout development. Journal of Cell Science.
Moving the nucleus to a specific position within the cell is an important event during many cell and developmental processes. Several different molecular mechanisms exist to position nuclei in various cell types. In this Commentary, we review the recent progress made in elucidating mechanisms of nuclear migration in a variety of important developmental models. Genetic approaches to identify mutations that disrupt nuclear migration in yeast, filamentous fungi, Caenorhabditis elegans, Drosophila and plants led to the identification of microtubule motors, as well as SUN and KASH proteins (LINC complex) that function to connect nuclei to the cytoskeleton. We focus on how these proteins and other mechanisms move nuclei throughout vertebrate development, including processes related to wound healing of fibroblasts, fertilization, developing myotubes and the developing central nervous system. We also describe how nuclear migration is involved in cells that migrate through constricted spaces. Based on these findings, it is becoming increasingly clear that defects in nuclear positioning are associated with human diseases.
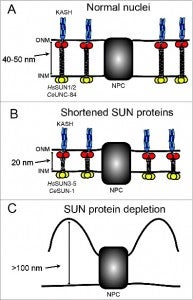
NE Cain, DA Starr (2014), SUN proteins and nuclear envelope spacing. Nucleus.
The nuclear envelope consists of 2 membranes separated by 30–50 nm, but how the 2 membranes are evenly spaced has been an open question in the field. Nuclear envelope bridges composed of inner nuclear membrane SUN proteins and outer nuclear membrane KASH proteins have been proposed to set and regulate nuclear envelope spacing. We tested this hypothesis directly by examining nuclear envelope spacing in Caenorhabditis elegans animals lacking UNC-84, the sole somatic SUN protein. SUN/KASH bridges are not required to maintain even nuclear envelope spacing in most tissues. However, UNC-84 is required for even spacing in body wall muscle nuclei. Shortening UNC-84 by 300 amino acids did not narrow the nuclear envelope space. While SUN proteins may play a role in maintaining nuclear envelope spacing in cells experiencing forces, our data suggest they are dispensable in most cells.
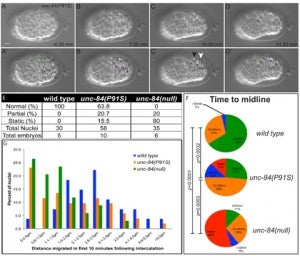
CR Bone, EC Tapley, M Gorjánácz, DA Starr (2014), The C. elegans SUN protein UNC-84 interacts with lamin to transfer forces from the cytoplasm to the nucleoskeleton during nuclear migration. Molecular Biology of the Cell.
Nuclear migration is a critical component of many cellular and developmental processes. The nuclear envelope forms a barrier between the cytoplasm, where mechanical forces are generated, and the nucleoskeleton. The LINC complex consists of KASH proteins in the outer nuclear membrane and SUN proteins in the inner nuclear membrane that bridge the nuclear envelope. How forces are transferred from the LINC complex to the nucleoskeleton is poorly understood. The C. elegans lamin, LMN-1, is required for nuclear migration and interacts with the nucleoplasmic domain of the SUN protein UNC-84. This interaction is weakened by the unc-84(P91S) missense mutation. These mutant nuclei have an intermediate nuclear migration defect—live imaging of nuclei or LMN-1::GFP show that many nuclei migrate normally, others initiate migration before subsequently failing, and others fail to begin migration. At least one other component of the nucleoskeleton, the NET5/Samp1/Ima1 homolog SAMP-1, plays a role in nuclear migration. We propose a nut and bolt model to explain how forces are dissipated across the nuclear envelope during nuclear migration. In this model, SUN/KASH bridges serve as bolts through the nuclear envelope, and nucleoskeleton components LMN-1 and SAMP-1 act as both nuts and washers on the inside of the nucleus.
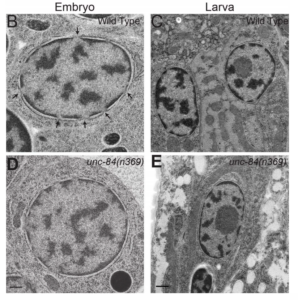
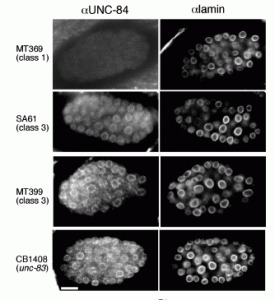
NE Cain, EC Tapley, KL McDonald, BM Cain, DA Starr (2014), The SUN protein UNC-84 is required only in force-bearing cells to maintain nuclear envelope architecture. The Journal of Cell Biology.
The nuclear envelope (NE) consists of two evenly spaced bilayers, the inner and outer nuclear membranes. The Sad1p and UNC-84 (SUN) proteins and Klarsicht, ANC-1, and Syne homology (KASH) proteins that interact to form LINC (linker of nucleoskeleton and cytoskeleton) complexes connecting the nucleoskeleton to the cytoskeleton have been implicated in maintaining NE spacing. Surprisingly, the NE morphology of most Caenorhabditis elegans nuclei was normal in the absence of functional SUN proteins. Distortions of the perinuclear space observed in unc-84 mutant muscle nuclei resembled those previously observed in HeLa cells, suggesting that SUN proteins are required to maintain NE architecture in cells under high mechanical strain. The UNC-84 protein with large deletions in its luminal domain was able to form functional NE bridges but had no observable effect on NE architecture. Therefore, SUN-KASH bridges are only required to maintain NE spacing in cells subjected to increased mechanical forces. Furthermore, SUN proteins do not dictate the width of the NE.
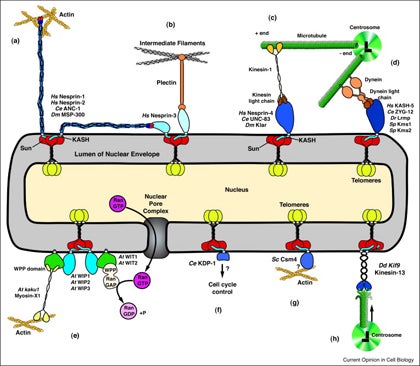
GWG Luxton, DA Starr (2014), KASHing up with the nucleus: novel functional roles of KASH proteins at the cytoplasmic surface of the nucleus. Current Opinion in Cell Biology.
Nuclear-cytoskeletal connections are central to fundamental cellular processes, including nuclear positioning and chromosome movements in meiosis. The cytoskeleton is coupled to the nucleoskeleton through conserved KASH-SUN bridges, or LINC complexes, that span the nuclear envelope. KASH proteins localize to the outer nuclear membrane where they connect the nucleus to the cytoskeleton. New findings have expanded the functional diversity of KASH proteins, showing that they interact with microtubule motors, actin, intermediate filaments, a nonconventional myosin, RanGAP, and each other. The role of KASH proteins in cellular mechanics is discussed. Genetic mutations in KASH proteins are associated with autism, hearing loss, cancer, muscular dystrophy and other diseases.
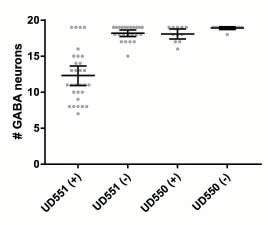
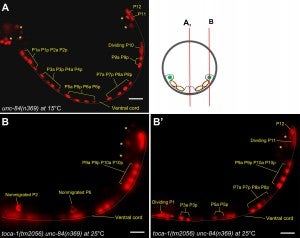
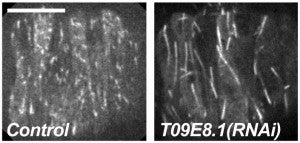
Y Chang, D Dranow, J Kuhn, M Meyerzon, M Ngo, D Ratner, K Warltier, DA Starr (2013), toca-1 is in a novel pathway that functions in parallel with a SUN-KASH nuclear envelope bridge to move nuclei in Caenorhabditis elegans. Genetics.
Moving the nucleus to an intracellular location is critical to many fundamental cell and developmental processes, including cell migration, differentiation, fertilization, and establishment of cellular polarity. Bridges of SUN and KASH proteins span the nuclear envelope and mediate many nuclear positioning events, but other pathways function independently through poorly characterized mechanisms. To identify and characterize novel mechanisms of nuclear migration, we conducted a non-biased forward genetic screen for mutations that enhanced the nuclear migration defect of unc-84, which encodes a SUN protein. In Caenorhabditis elegans larvae, failure of hypodermal P-cell nuclear migration results in uncoordinated and egg-laying defective animals. The process of P-cell nuclear migration in unc-84 null animals is temperature sensitive; at 25°C migration fails in unc-84mutants, but at 15°C the migration occurs normally. We hypothesized that an additional pathway functions in parallel to the unc-84 pathway to move P-cell nuclei at 15°C. In support of our hypothesis, forward genetic screens isolated eight emu (for enhancer of the nuclear migration defect of unc-84) mutations that only disrupt nuclear migration in a null unc-84 background. The yc20 mutant was determined to carry a mutation in the toca-1 gene. TOCA-1 functions to move P-cell nuclei in a cell-autonomous manner. TOCA-1 is conserved in humans, where it functions to nucleate and organize actin during endocytosis. Therefore, we have uncovered a player in a previously unknown, likely actin-dependent, pathway that functions to move nuclei in parallel to SUN-KASH bridges. The other emumutations potentially represent other components of this novel pathway.
EC Tapley, DA Starr (2012), Connecting the nucleus to the cytoskeleton by SUN–KASH bridges across the nuclear envelope. Current Opinion in Cell Biology.
The nuclear–cytoskeleton connection influences many aspects of cellular architecture, including nuclear positioning, the stiffness of the global cytoskeleton, and mechanotransduction. Central to all of these processes is the assembly and function of conserved SUN–KASH bridges, or LINC complexes, that span the nuclear envelope. Recent studies provide details of the higher order assembly and targeting of SUN proteins to the inner nuclear membrane. Structural studies characterize SUN–KASH interactions that form the central link of the nuclear-envelope bridge. KASH proteins at the outer nuclear membrane link the nuclear envelope to the cytoskeleton where forces are generated to move nuclei. Significantly, SUN proteins were recently shown to contribute to the progression of laminopathies.
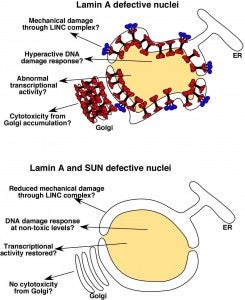
DA Starr (2012), Laminopathies: too much SUN is a bad thing. Current Biology.
SUN proteins accelerate the pathological progression of laminopathies. Although the mechanisms of how this occurs remain to be elucidated, an intriguing possibility is that high levels of Sun proteins lead to a hyperactive DNA damage response.
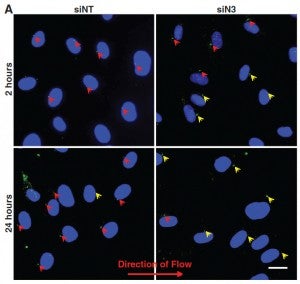
JT Morgan, ER Pfeiffer, TL Thirkill, P Kumar, G Peng, HN Fridolfsson, GC Douglas, DA Starr*, AI Barakat* (2011), Nesprin-3 regulates endothelial cell morphology, perinuclear cytoskeletal architecture, and flow-induced polarization. Molecular Biology of the Cell. (*Equal Contribution)
Changes in blood flow regulate gene expression and protein synthesis in vascular endothelial cells, and this regulation is involved in the development of atherosclerosis. How mechanical stimuli are transmitted from the endothelial luminal surface to the nucleus is incompletely understood. The linker of nucleus and cytoskeleton (LINC) complexes have been proposed as part of a continuous physical link between the plasma membrane and subnuclear structures. LINC proteins nesprin-1, -2, and -4 have been shown to mediate nuclear positioning via microtubule motors and actin. Although nesprin-3 connects intermediate filaments to the nucleus, no functional consequences of nesprin-3 mutations on cellular processes have been described. Here we show that nesprin-3 is robustly expressed in human aortic endothelial cells (HAECs) and localizes to the nuclear envelope. Nesprin-3 regulates HAEC morphology, with nesprin-3 knockdown inducing prominent cellular elongation. Nesprin-3 also organizes perinuclear cytoskeletal organization and is required to attach the centrosome to the nuclear envelope. Finally, nesprin-3 is required for flow-induced polarization of the centrosome and flow-induced migration in HAECs. These results represent the most complete description to date of nesprin-3 function and suggest that nesprin-3 regulates vascular endothelial cell shape, perinuclear cytoskeletal architecture, and important aspects of flow-mediated mechanotransduction.
EC Tapley, N Ly, DA Starr (2011), Multiple mechanisms actively target the SUN protein UNC-84 to the inner nuclear membrane. Molecular Biology of the Cell.
Approximately 100 proteins are targeted to the inner nuclear membrane where they regulate chromatin and nuclear dynamics. However, the mechanisms underlying trafficking to the inner nuclear membrane are poorly understood. The Caenorhabditis elegans SUN protein UNC-84 is an excellent model to investigate such mechanisms. UNC-84 recruits KASH proteins to the outer nuclear membrane to bridge the nuclear envelope, mediating nuclear positioning. UNC-84 has four targeting sequences: two classical nuclear localization signals, an inner nuclear membrane sorting motif, and a signal conserved in mammalian Sun1, the SUN-nuclear-envelope-localization signal. Mutations in some signals disrupt the timing of UNC-84 nuclear-envelope localization, showing that diffusion is not sufficient to move all UNC-84 to the nuclear envelope. Thus, targeting UNC-84 requires an initial step that actively transports UNC-84 from the peripheral ER to the nuclear envelope. Only when all four signals are simultaneously disrupted does UNC-84 completely fail to localize and to function in nuclear migration, meaning that at least three signals function, in part, redundantly to ensure proper targeting of UNC-84. Multiple mechanisms might also be used to target other proteins to the inner nuclear membrane, thereby ensuring their proper and timely localization for essential cellular and developmental functions.
RA Green, HL Kao, A Audhya, S Arur, JR Mayers, HN Fridolfsson, M Schulman, S Schloissnig, S Niessen, K Laband, S Wang, DA Starr, AA Hyman, T Schedl, A Desai, F Piano, KC Gunsalus, K Oegema (2011), A high-resolution C. elegans essential gene network based on phenotypic profiling of a complex tissue. Cell.
High-content screening for gene profiling has generally been limited to single cells. Here, we explore an alternative approach-profiling gene function by analyzing effects of gene knockdowns on the architecture of a complex tissue in a multicellular organism. We profile 554 essential C. elegans genes by imaging gonad architecture and scoring 94 phenotypic features. To generate a reference for evaluating methods for network construction, genes were manually partitioned into 102 phenotypic classes, predicting functions for uncharacterized genes across diverse cellular processes. Using this classification as a benchmark, we developed a robust computational method for constructing gene networks from high-content profiles based on a network context-dependent measure that ranks the significance of links between genes. Our analysis reveals that multi-parametric profiling in a complex tissue yields functional maps with a resolution similar to genetic interaction-based profiling in unicellular eukaryotes-pinpointing subunits of macromolecular complexes and components functioning in common cellular processes.
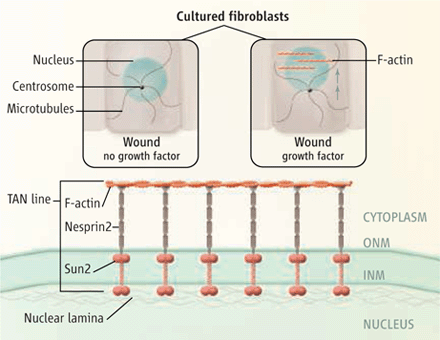
DA Starr (2010), Nuclei get TAN lines. Science invited perspective.
Positioning the nucleus within a cell involves a protein complex that spans the nuclear envelope to connect actin filaments to the nuclear lamina.
A perspective of Luxton et al. 2010. “Linear arrays of nuclear envelope proteins harness retrograde actin flow for nuclear movement.” Science 20; 956-959
DA Starr, HN Fridolfsson (2010), Interactions between Nuclei and the Cytoskeleton are Mediated by SUN-KASH Nuclear-Envelope Bridges. Annual Review of Cell and Developmental Biology.
The nuclear envelope links the cytoskeleton to structural components of the nucleus. It functions to coordinate nuclear migration and anchorage, organize chromatin, and aid meiotic chromosome pairing. Forces generated by the cytoskeleton are transferred across the nuclear envelope to the nuclear lamina through a nuclear-envelope bridge consisting of SUN (Sad1 and UNC-84) and KASH proteins (Klarsicht, ANC-1 and Syne/Nesprin homology). Some KASH-SUN combinations connect microtubules, centrosomes, actin filaments, or intermediate filaments to the surface of the nucleus. Other combinations are used in cell cycle control, nuclear import, or apoptosis. Interactions between the cytoskeleton and the nucleus also affect global cytoskeleton organization. SUN and KASH proteins were identified through genetic screens for mispositioned nuclei in model organisms. Knockouts of SUN or KASH proteins disrupt neurological and muscular development in mice. Defects in SUN and KASH proteins have been linked to human diseases including muscular dystrophy, ataxia, progeria, lissencephaly, and cancer.
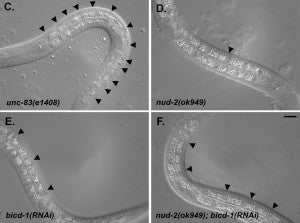
HN Fridolfsson, N Ly, M Meyerzon, DA Starr (2010), UNC-83 coordinates kinesin-1 and dynein activities at the nuclear envelope during nuclear migration. Developmental Biology.
Nuclei migrate during many events, including fertilization, establishment of polarity, differentiation, and cell division. The C. elegans KASH protein UNC-83 localizes to the outer nuclear membrane where it recruits kinesin-1 to provide the major motor activity required for nuclear migration in embryonic hyp7 cells. Here we show that UNC-83 also recruits two dynein-regulating complexes to the cytoplasmic face of the nucleus that play a regulatory role. One consists of the NudE homolog NUD-2 and the NudF/Lis1/Pac1 homolog LIS-1; the other includes dynein light chain DLC-1, the BicaudalD homolog BICD-1, and the egalitarian homologue EGAL-1. Genetic disruption of any member of these two complexes caused nuclear migration defects that were enhanced in some double mutant animals, suggesting that BICD-1 and EGAL-1 function in parallel to NUD-2. Dynein heavy chain mutant animals also had a nuclear migration defect, suggesting these complexes function through dynein. Deletion analysis indicated that independent domains of UNC-83 interact with kinesin and dynein. These data suggest a model where UNC-83 acts as the cargo-specific adaptor between the outer nuclear membrane and the microtubule motors kinesin-1 and dynein. Kinesin-1 functions as the major force generator during nuclear migration, while dynein is involved in regulation of bidirectional transport of the nucleus.
MD McGee, I Stagljar, DA Starr (2009), KDP-1 is a nuclear envelope KASH protein required for cell cycle progression. Journal of Cell Science.
KASH proteins localize to the outer nuclear membrane where they connect the nucleus to the cytoskeleton. KASH proteins interact with SUN proteins to transfer forces across the nuclear envelope to position nuclei or move chromosomes. A new KASH protein, KDP-1, was identified in a membrane yeast two-hybrid screen of a C. elegans library using the SUN protein UNC-84 as bait. KDP-1 also interacted with SUN-1. KDP-1 was enriched at the nuclear envelope in a variety of tissues and required SUN-1 for nuclear envelope localization in the germline. Genetic analyses showed that kdp-1 was essential for embryonic viability, larval growth, and germline development. kdp-1(RNAi) delayed the entry into mitosis in embryos, led to a small mitotic zone in the germline, and caused an endomitotic phenotype. Aspects of these phenotypes were similar to those seen in sun-1(RNAi) , suggesting that KDP-1 functions with SUN-1 in the germline and early embryo. The data suggest that KDP-1 is a novel KASH protein that functions to ensure the timely progression of the cell cycle between the end of S phase and the entry into mitosis.
M Meyerzon, HN Fridolfsson, N Ly, FJ McNally, DA Starr (2009), UNC-83 is a nuclear-specific cargo adaptor for kinesin-1 mediated nuclear migration. Development.
Intracellular nuclear migration is essential for many cellular events including fertilization, establishment of polarity, division, and differentiation. How nuclei migrate is not understood at the molecular level. The C. elegans KASH protein UNC-83 is required for nuclear migration and localizes to the outer nuclear membrane. UNC-83 interacts with the inner nuclear membrane SUN protein, UNC-84, and is proposed to connect the cytoskeleton to the nuclear lamina. Here, we show that UNC-83 also interacts with the kinesin-1 light chain KLC-2 as identified in a yeast two-hybrid screen and confirmed by in vitro assays. UNC-83 interacts with and recruits KLC-2 to the nuclear envelope in a heterologous tissue culture system. Additionally, analysis of mutant phenotypes demonstrated that both KLC-2 and the kinesin-1 heavy chain, UNC-116, are required for nuclear migration. Finally, we can partially bypass the requirement for UNC-83 in nuclear migration by expressing a synthetic outer nuclear membrane KLC-2::KASH fusion protein. Our data support a model where UNC-83 plays a central role in nuclear migration by acting to bridge the nuclear envelope and as a kinesin-1 cargo-specific adaptor so that motor-generated forces specifically move the nucleus as a single unit.
DA Starr (2009), A nuclear-envelope bridge positions nuclei and moves chromosomes. Journal of Cell Science.
Positioning the nucleus is essential for the formation of polarized cells, pronuclear migration, cell division, cell migration and the organization of specialized syncytia such as mammalian skeletal muscles. Proteins that are required for nuclear positioning also function during chromosome movements and pairing in meiosis. Defects in these processes lead to human diseases including laminopathies. To properly position the nucleus or move chromosomes within the nucleus, the cell must specify the outer surface of the nucleus and transfer forces across both membranes of the nuclear envelope. KASH proteins are specifically recruited to the outer nuclear membrane by SUN proteins, which reside in the inner nuclear membrane. KASH and SUN proteins physically interact in the perinuclear space, forming a bridge across the two membranes of the nuclear envelope. The divergent N-terminal domains of KASH proteins extend from the surface of the nucleus into the cytoplasm and interact with the cytoskeleton while the N termini of SUN proteins extend into the nucleoplasm to interact with the lamina or chromatin. The SUN and KASH bridge across the nuclear envelope functions to transfer forces generated in the cytoplasm into the nucleoplasm during nuclear migration, nuclear anchorage, centrosome attachment, intermediate-filament association and telomere clustering.
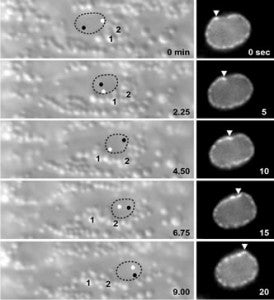
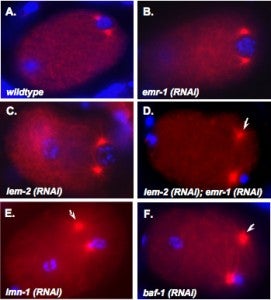
M Meyerzon, Z Gao, J Liu, J Wu, CJ Malone, DA Starr (2009), Centrosome attachment to the C. elegans male pronucleus is dependent on the surface area of the nuclear envelope. Developmental Biology.
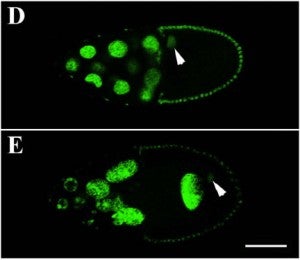
A close association must be maintained between the male pronucleus and the centrosomes during pronuclear migration. In C. elegans, simultaneous depletion of inner nuclear membrane LEM proteins EMR-1 and LEM-2, depletion of the nuclear lamina proteins LMN-1 or BAF-1, or the depletion of nuclear import components leads to embryonic lethality with small pronuclei. Here, a novel centrosome detachment phenotype in C. elegans zygotes is described. Zygotes with defects in the nuclear envelope had small pronuclei with a single centrosome detached from the male pronucleus. ZYG-12, SUN-1, and LIS-1, which function at the nuclear envelope with dynein to attach centrosomes, were observed at normal concentrations on the nuclear envelope of pronuclei with detached centrosomes. Analysis of time-lapse images showed that as mutant pronuclei grew in surface area, they captured detached centrosomes. Larger tetraploid or smaller histone::mCherry pronuclei suppressed or enhanced the centrosome detachment phenotype respectively. In embryos fertilized with anucleated sperm, only one centrosome was captured by small female pronuclei, suggesting the mechanism of capture is dependent on the surface area of the outer nuclear membrane available to interact with aster microtubules. We propose that the limiting factor for centrosome attachment to the surface of abnormally small pronuclei is dynein.
DA Starr (2007), Communication between the cytoskeleton and the nuclear envelope to position the nucleus. Molecular BioSystems.
In most eukaryotic cells, the nucleus is localized to a specific location. This highlight article focuses on recent advances describing the mechanisms of nuclear migration and anchorage. Central to nuclear positioning mechanisms is the communication between the nuclear envelope and the cytoskeleton. All three components of the cytoskeleton, microtubules, actin filaments and intermediate filaments, are involved in nuclear positioning to varying degrees in different cell types. KASH proteins on the outer nuclear membrane connect to SUN proteins on the inner nuclear membrane. Together they transfer forces between the cytoskeleton and the nuclear lamina. Once at the outer nuclear membrane, KASH proteins can interact with the cytoskeleton. Nuclear migrations are a component of many cellular migration events and defects in nuclear positioning lead to human diseases, most notably lissencephaly.
MD McGee*, R Rillo*, AS Anderson, DA Starr (2006), UNC-83 is a KASH protein required for nuclear migration and is recruited to the outer nuclear membrane by a physical interaction with the SUN protein UNC-84. Molecular Biology of the Cell. (*Equal Contribution)
UNC-84 is required to localize UNC-83 to the nuclear envelope where it functions during nuclear migration. A KASH domain in UNC-83 was identified. KASH domains are conserved in the nuclear envelope proteins Syne/nesprins, Klarsicht, MSP-300, and ANC-1. C. elegans UNC-83 was shown to localize to the outer nuclear membrane and UNC-84 to the inner nuclear membrane in transfected mammalian cells, suggesting the KASH and SUN protein targeting mechanisms are conserved. Deletion of the KASH domain of UNC-83 blocked nuclear migration and localization to the C. elegans nuclear envelope. Some point mutations in the UNC-83 KASH domain disrupted nuclear migration, even if they localized normally. At least two separable portions of the C-terminal half of UNC-84 were found to interact with the UNC-83 KASH domain in a membrane-bound, split-ubiquitin yeast two-hybrid system. However, the SUN domain was essential for UNC-84 function and UNC-83 loclization in vivo. These data support the model that KASH and SUN proteins bridge the nuclear envelope, connecting the nuclear lamina to cytoskeletal components. This mechanism appears conserved across eukaryotes and is the first proposed mechanism to target proteins specifically to the outer nuclear membrane.
J Yu, DA Starr, X Wu, SM Parkhurst, Y Zhuang, T Xu, R Xu, M Han (2006), The KASH domain protein MSP-300 plays an essential role in nuclear anchoring during Drosophila oogenesis. Developmental Biology.
During late stages of Drosophila oogenesis, the cytoplasm of nurse cells in the egg chamber is rapidly transferred (‘‘dumped’’) to oocytes, while the nurse cell nuclei are anchored by a mechanism that involves the actin cytoskeleton. The factors that mediate this interaction between nuclei and actin cytoskeleton are unknown. MSP-300 is the likely Drosophila ortholog of the mammalian Syne-1 and -2 and C. elegans ANC-1 proteins, contained both actin-binding and nuclear envelope localization domains. By using an antibody against C-terminus of MSP-300, we find that MSP-300 is distributed throughout the cytoplasm and accumulates at the nuclear envelope of nurse cells and the oocyte. A GFP fusion protein containing the C-terminal region of MSP-300 is also sufficient to localize protein on the nuclear envelope in oocytes. To eliminate the maternal gene activity during oogenesis, we generated homozygous germ-line clones of a loss-of-function mutation in msp-300 in otherwise heterozygous mothers. In the mutant egg chambers that develop from such clones, cytoplasmic dumping of nurse cells is severely disturbed. The nuclei of nurse cells and the oocyte are mislocalized and the usually well-organized actin structures are severely disrupted. These results indicate that maternal MSP-300 plays an important role in actin-dependent nuclear anchorage during cytoplasmic transport.
DA Starr, JA Fischer (2005), KASH ‘n Karry: the KASH domain family of cargo-specific cytoskeletal adaptor proteins. Bioessays.
A diverse family of proteins has been discovered with a small C-terminal KASH domain in common. KASH domain proteins are localized uniquely to the outer nuclear envelope, enabling their cytoplasmic extensions to tether the nucleus to actin filaments or microtubules. KASH domains are targeted to the outer nuclear envelope by SUN domains of inner nuclear envelope proteins. Several KASH protein genes were discovered as mutant alleles in model organisms with defects in developmentally regulated nuclear positioning. Recently, KASH-less isoforms have been found that connect the cytoskeleton to organelles other than the nucleus. A widened view of these proteins is now emerging, where KASH proteins and their KASH-less counterparts are cargo-specific adaptors that not only link organelles to the cytoskeleton but also regulate developmentally specific organelle movements.
MA Ruegg (2005), Organization of synaptic myonuclei by Syne proteins and their role during the formation of the nerve–muscle synapse. Commentary in
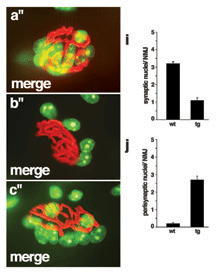
Vertebrate skeletal muscle fibers contain hundreds of nuclei, of which three to six are functionally specialized and stably anchored beneath the postsynaptic membrane at the neuromuscular junction (NMJ). The mechanisms that localize synaptic nuclei and the roles they play in neuromuscular development are unknown. Syne-1 is concentrated at the nuclear envelope of synaptic nuclei; its Caenorhabditis elegans orthologue ANC-1 functions to tether nuclei to the cytoskeleton. To test the involvement of Syne proteins in nuclear anchoring, we generated transgenic mice overexpressing the conserved C-terminal Klarsicht ANC-1 Syne homology domain of Syne-1. The transgene acted in a dominant interfering fashion, displacing endogenous Syne-1 from the nuclear envelope. Muscle nuclei failed to aggregate at the NMJ in transgenic mice, demonstrating that localization and positioning of synaptic nuclei require Syne proteins. We then exploited this phenotype to show that synaptic nuclear aggregates are dispensable for maturation of the NMJ.
DA Starr, M Han (2005), A genetic approach to study the role of nuclear envelope components in nuclear positioning. Nuclear organization in development and disease, Wiley (Novartis Found Symp 264).
In many cell types, the nucleus is positioned to a specific location. Our work and others have demonstrated that several integral nuclear envelope proteins function to move the nucleus and to anchor it in place. Our forward genetic approach has identified three components of the nuclear envelope involved in nuclear positioning. ANC-1 consists of two actin-binding calponin domains, a huge central coiled domain, and a nuclear envelope targeting domain termed the KASH domain. ANC-1 functions to physically tether the actin cytoskeleton to the outer nuclear membrane. UNC-83 is a novel protein that functions in an unknown manner during nuclear migration. UNC-83 contains a domain with weak homology to the KASH domain of ANC-1. UNC-84 is a SUN protein that is required for both nuclear migration and anchorage. UNC-84 recruits both UNC-83 and ANC-1 to the nuclear envelope. We propose a model where UNC-84 is an integral component of the inner nuclear membrane, with its SUN domain in the perinuclear space. The SUN domain then recruits ANC-1 and UNC-83, through interactions with their KASH domains, to the outer nuclear envelope. Together these proteins function to bridge the two membranes of the nuclear envelope, connecting the nuclear matrix to the cytoskeleton.
DA Starr, M Han (2003), ANChors away: an actin based mechanism of nuclear positioning. Journal of Cell Science.
Mechanisms for nuclear migration and nuclear anchorage function together to control nuclear positioning. Both tubulin and actin networks play important roles in nuclear positioning. The actin cytoskeleton has been shown to position nuclei in a variety of systems from yeast to plants and animals. It can either act as a stable skeleton to anchor nuclei or supply the active force to move nuclei. Two C. elegans genes and their homologues play important roles in these processes. Syne/ANC-1 anchors nuclei by directly tethering the nuclear envelope to the actin cytoskeleton, and UNC-84/SUN functions at the nuclear envelope to recruit Syne/ANC-1.
DA Starr, M Han (2002), Role of ANC-1 in tethering nuclei to the actin cytoskeleton. [Supplement] Science.
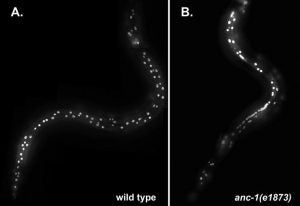
Mutations in anc-1 (nuclear anchorage defective) disrupt the positioning of nuclei and mitochondria in Caenorhabditis elegans. ANC-1 is shown to consist of mostly coiled regions with a nuclear envelope localization domain (called the KASH domain) and an actin-binding domain; this structure was conserved with the Drosophila protein Msp-300 and the mammalian Syne proteins. Antibodies against ANC-1 localized cytoplasmically and were enriched at the nuclear periphery in an UNC-84–dependent manner. Overexpression of the KASH domain or the actin-binding domain caused a dominant negative anchorage defect. Thus, ANC-1 may connect nuclei to the cytoskeleton by interacting with UNC-84 at the nuclear envelope and with actin in the cytoplasm.
KK Lee*, DA Starr*, M Cohen, J Liu, M Han, KL Wilson, Y Gruenbaum (2002), Lamin-dependent localization of UNC-84, a protein required for nuclear migration in C. elegans. Molecular Biology of the Cell. (*Equal Contribution)
Mutations in the Caenorhabditis elegans unc-84 gene cause defects in nuclear migration and anchoring. We show that endogenous UNC-84 protein colocalizes with Ce-lamin at the nuclear envelope and that the envelope localization of UNC-84 requires Ce-lamin. We also show that during mitosis, UNC-84 remains at the nuclear periphery until late anaphase, similar to known inner nuclear membrane proteins. UNC-84 protein is first detected at the 26-cell stage and thereafter is present in most cells during development and in adults. UNC-84 is properly expressed in unc-83 and anc-1 lines, which have phenotypes similar to unc-84, suggesting that neither the expression nor nuclear envelope localization of UNC-84 depends on UNC-83 or ANC-1 proteins. The envelope localization of Ce-lamin, Ce-emerin, Ce-MAN1, and nucleoporins are unaffected by the loss of UNC-84. UNC-84 is not required for centrosome attachment to the nucleus because centrosomes are localized normally in unc-84 hyp7 cells despite a nuclear migration defect. Models for UNC-84 localization are discussed.
DS Fay, DA Starr, A Spencer, W Johnson. Edited by A Fluet, J Yochem (2001), Worm Breeding for Super Geniuses: A guide to genetic mapping in C. elegans.
What this guide is and isn’t. This guide is neither a basic text in Mendelian genetics nor is it in any way a comprehensive description of C. elegans biology. Detailed information about the latter can be gleaned from C. elegans I and C. elegans II by Cold Spring Harbor Press Inc. This guide does assume a working knowledge of basic genetics and will be of limited use to those who lack some background in this area. It is our hope that this text may serve as a supplement to existing published materials and that it will facilitate the successful breeding of worms by those new to the field.
DA Starr, GJ Hermann, CJ Malone, W Fixsen, JR Priess, HR Horvitz, M Han (2001), unc-83 encodes a novel component of the nuclear envelope and is essential for proper nuclear migration. Development.
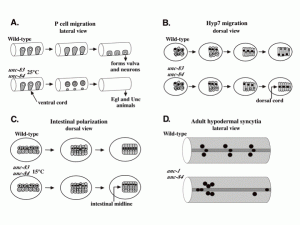
Nuclear migration plays an essential role in the growth and development of a wide variety of eukaryotes. Mutations in unc-84, which encodes a conserved component of the nuclear envelope, have been shown to disrupt nuclear migration in two C. elegans tissues. We show that mutations in unc-83 disrupt nuclear migration in a similar manner in migrating P cells, hyp7 precursors and the intestinal primordium, but have no obvious defects in the association of centrosomes with nuclei or the structure of the nuclear lamina of migrating nuclei. We also show that unc-83 encodes a novel transmembrane protein. We identified three unc-83 transcripts that are expressed in a tissuespecific manner. Antibodies against UNC-83 co-localized to the nuclear envelope with lamin and UNC-84. Unlike UNC- 84, UNC-83 localized to only specific nuclei, many of which were migratory. UNC-83 failed to localize to the nuclear envelope in unc-84 mutants with lesions in the conserved SUN domain of UNC-84, and UNC-83 interacted with the SUN domain of UNC-84 in vitro, suggesting that these two proteins function together during nuclear migration. We favor a model in which UNC-84 directly recruits UNC-83 to the nuclear envelope where they help transfer force between the cytoskeleton and the nucleus.